In last week's Part 1, I gave an introduction to the problem of categorizing the polarity of morphological traits. How can we reconstruct which characters are primitive, or plesiomorphic according to Hennig, and which are derived, or apomorphic? This is something we need to do to reconstruct evolution, because most of the past is only preserved in the form of fossils, usually lacking any DNA. In this second part of the discussion, I'm going to take apart my own tree and show why we inevitably need networks, not trees.
There may be more than one tree
Even with more and more data at hand, some molecular phylogenies refuse to be unambiguous. Even worse, different, well-sampled molecular data sets may tell different stories — ie. there is more than one molecular tree to explain the diversity patterns. The ML tree used for the ML character mapping in Part 1 was pretty well supported, but not telling the entire truth.
For a start, there is no reason to assume that oaks are not monophyletic even though the data fail to resolve them as a clade (evolving something unique like the oaks twice would be a striking trick, even for gambling Mother Nature) — molecular trees may have misleading, sometimes just wrong, branches, even when they are highly supported.
In this case, one complication is that the oligogene dataset combines plastid and nuclear gene regions that not only differ in their information content but also infer different phylogenetic scenarios (and mask a lot of intra-generic and sub-generic incongruencies). This is illustrated in the following tanglegram.
Even though the support along the backbone of the plastid tree is lowa (to non-existent), it well reflects the general diversification patterns in Fagaceae plastomes (see also the tree in Manos et al. 2008, Madroño 55:181–190; and Yan et al. 2019, BMC Evol. Biol. 19: 202, for an oak global picture). Plastid signatures show a strong geographic sorting (eg. New World vs. Old World), while the nuclear data provides most of the lineage-differentiating signal expressed in the combined tree (Part 1, Fig. 2).
Mapping along networks
How do we decide what is a real synapomorphy, a homoiology, or a good symplesiomorphy? Mapping the traits along all possible rooted trees is one option. Another option is to just map them along a consensus network of all trees, as shown next.
According to the mapping, the newly described South American Castanopsis rothwellii, assigned to the modern (Souteast Asian) genus Castanopsis, is a stem Castanoideae / Fagaceae, while the "extinct" North American genus Castanopsoidea (then the "earliest megafossil evidence of Fagaceae": Crepet & Nixon 1989, Am. J. Bot. 76: 842–855) could be a stem / crown member of the Castanea-Castanopsis lineage. The difference to the ML trait mapping (Fig. 3 in Part 1) on the combined tree is that we get a better picture what is a lineage-specific trait set in Castanea-Castanopsis, because the interference of the monophyletic(!) oak grade is minimized.
Another possibility is to map the characters directly along a distance-based network, and then compare the latter with the molecular-based topological alternatives. This is quite puzzling in this case, because the morphology (Fig. 1 in Part 1) matches neither the nuclear tree nor the plastid tree (Figs. 2–4) — the traits scored for the fossils cover largely morphological Play-Doh of the Fagaceae.
The likely primitive characters, irrespective of the evolutionary scenario we prefer, are those also found in the Eocene fossilsb. There are no derived traits/character suites pinning the fossils to Castanopsis. The fossils are a bit derived on their own terms (note their position in Fig. 5), and hence we can deduce that the fossils are either: (a) representing a relatively primitive extinct American sister lineage or (b) surviving, somewhat evolved members of the precursors of modern-day core Fagaceae. Note that the derived oaks evolved nearly 60 myrs ago, ie. 8 myrs before the oldest (Patagonian) Castanoideae fossil was deposited. The earliest (known) Fagaceae and castaneoid pollen are from 80+ Ma old Upper Cretaceous sediments in western North America (Grímsson et al. 2016, Acta Palaeobot. 56: 247–305; open access) and Japan (Takahashi et al. 2008, Intl. J. Plant Sci. 169:899–907), giving them plenty of time to migrate into North and then South America during the Paleocene-Eocene green house episode.
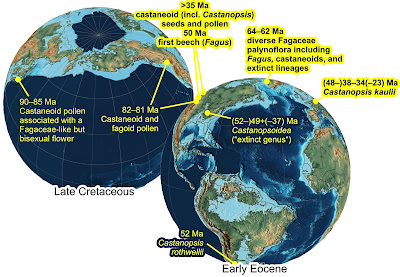
|
| Fig. 6 – Earliest fossil record of Fagaceae and Castanoideae mapped on Scotese's Paleoglobes (© Scotese 2013, GoogleEarth layover files are available from here). Note that although there was no continuous land bridge, North and South America were already connected by a chain of large and high islands, providing a corridor for intercontinental dispersal of near- and extra-tropical plant lineages. A potential crown-group Castanopsis (C. kaulii, cupule with associated seeds and pollen) has been recently recovered from the Baltic Amber (Sadowski et al. 2018 Am. J. Bot 105: 2025–2036). |
Both of the mapping procedures described above are crude, in the sense that they ignore the molecular branch lengths, and use Ockham's Razor. But it strikes me as being not a bad start. They are better than just mapping along a single preferred molecular tree (as is done in many neontological papers; see Part 1) or along a morphology-based strict consensus cladogram (as is done in far too many paleontological papers; many palaeobotanical papers do neither the one nor the other: eg. Wilf et al., 2019, Science 364: eaaw5139). It's important to realize that if one taxon or subtree of our modern taxon set is characterized solely by the lack of shared derived traits or unstable expression of derived traits (like Castanopsis here, see position in both graphs in Fig. 5), ie. represents living fossils or little-evolved lineages, any ancient and primitive fossil, stem group, sister group or precursor, will be attracted by them in a total evidence or any other tree-based approach, especially when we rely on change-probability-naive parsimony as inference criterion. As we pointed out repeatedly: forming a clade in tree is neither a necessary nor a sufficient criterion for monophyly.
All gone, what to do when we have no molecular data?
Morphology alone, like genes on their own, will inevitably get some things wrong (compare Fig. 4 with Fig. 5). Without molecular data, one may have little reason to reject the monophyly of the Castaneoideae (when using more than the seven characters scored by Wilf et al. 2019; see eg. the cladogram in Crepet & Nixon 1989, fig. 1 based on an undocumented 25-character matrix). In the process, we would misinterpret overall similarity, due to shared primitive character suites and the lack of shared derived traits as evidence for an inclusive common originc.
What can we do if we have no or very few extant taxa, when we only have one set of data prone to circular reasoning? Then using networks is inevitable as well (see Fig. 5; and some examples provided in the reading list below). We need to explore in-depth the signal in our data matrix. Only extremely biased morphological matrices provide clear tree-like signals, comprehensive ones will have internal conflict and allow for inferring many, partly very different but more or less equally optimal trees.
Exploratory data analysis will not eliminate all possible errors — based only on the graph in Fig. 5, we would get the inter-generic phylogenetic relationships in Fagaceae partly wrong. However, this may lead to an informed decision as to which of the many equally probable evolutionary scenarios make more sense than others. It will help to reduce the alternatives, without eliminating those that are equally valid (which every tree does). If the time-coverage is good, exploring morphological differentiation over time can be an asset, too (see eg. Stacking neighbor-nets – a real-world example).
Data
The matrices used, networks etc. can be accessed via figshare.
Selection of related posts on The Genealogical World of Phylogenetic Networks
Clades, Cladograms, Cladistics, and why networks are inevitable — illustrates why paleontologists should also be less tree-naive (see example in footnote c).
Has homoiology be neglected in phylogenetics? — why we should try to assess the phylogenetic quality of our traits.
Let distinguish between Hennig and Cladistics — as said in the title, the post provides reasons why we should distinguish between Hennig's concepts and clades in phylogenetic trees.
Ockham's Razor applied, but not used: can we do DNA-scaffolding with seven characters? — the original post dealing with Wilf et al.'s (2019) "phylogenetic analysis", which obviously was not scrutinized during review.
Please stop use cladograms! — No matter whether you think evolution is tree-like or not, cladograms should be a matter of the past.
Should we try to infer trees on tree-unlikely matrices? — using well-known (among paleobotanists) examples, I show why networks reveal much more than any tree when we deal with fossils.
More non-treelike data forced into trees: a glimpse into the dinosaurs — the same but for a thunder lizard matrix.
Trivial data, but not so trivial graphs — an inference experiment using very simple artificial binary matrices.
a The main reason for the lack of branch support is that individuals of different genera growing in the same area can share plastid haplotypes, while individuals of the same genus / infra-generic lineage, even species, can be quite different. [Note that the standard 4x4 ML nucleotide model treats polymorphisms as such, not as missing data.] Plus, the different lineages show different levels of plastid diversity (highest in Quercus subgenus Cerris, but low in subgenus Quercus, the North American castanoids and Lithocarpus outside Borneo, Castanea-Castanopsis appear to be in-between the extremes), and there is a tendency to preferably mutate sequence patterns within a lineage that otherwise differentiate between lineages (for instance, inversions that distinguish two genera, can be found as intra-lineage variation in the third genus or one of the oak sections).
b The striking similarity between the newly found South American and long-known slightly older North American fossils is likely the reason for not discussing the latter in the original paper or including them in the "DNA-scaffold" analysis. As is obvious from the graphs, the slightly younger North American fossil could easily be a slightly more derived of the same lineage than the South American fossil (Planchard et al. 2016 Paleont. Electr. 19.3.51A give a revised age of ≥ 49 Ma for the plant-bearing strata), and thus would have been at odds with the narrative of the authors (see also comment by Denk et al. 2019, Science 10.1126/science.aaz2189).
c As done by Wilf et al. (see also the argumentation in Wilf et al.'s response, Science 10.1126/science.aaz2297, to Denk et al.'s 2019 comment). The combination of circular reasoning, systematic bias, and (parsimony) tree-naivity is well expressed in Wilf et al.'s own words:
Fourth, Denk et al. erroneously contend that Castanopsis rothwellii, a fossil with so many diagnostic characters preserved that it could only be assigned to Castanopsis if “found alive” today (1), has plesiomorphic features and cannot be placed confidently in the extant genus [see Figs. 1–5 in this two-part post]. ... Denk et al.’s phylogenetic conclusions from their emended tree and matrix are misleading, in that any morphological matrix includes characters that are relevant only for the taxa included in the analysis. ... Because the fossils are castaneoid in all features, we did not include all Fagaceae in our original analysis (1) and likewise did not include all characters relevant to non-castaneoid fagaceous taxa. ... By adding just three relevant characters to the Denk et al. scaffold to accommodate the genera they added (Table 1), the fossil Castanopsis rothwellii is placed only with Castanopsis in the single [ie. the strict consensus of two equally parsimonious trees] most parsimonious tree (Fig. 1).One of the three added traits ("expanded stigma") is exclusively shared by all five Castaneoideae genera, the second ("nut generally rounded in cross section") shared by all but one Castaneoideae and Quercus, and thus are symplesiomorphies of core Fagaceae: shared primitive traits that can be expected in a precursor of several or all modern genera or their less evolved extinct sister lineages. Or positively selected homoiologies, ie. evolved multiple times within the core Fagaceae. The third ("asymmetrical cupule") is an unstable convergence / deep parallelism and a trait of little phylogenetic value, since expressed as intra-generic (intraspecific?) variation in two distantly related genera: the monotypic Formanodendron, a trigonobalanoid, and Castanopsis. These are two genera that share only a very distant (and exclusive fide Hennig) common origin (see Part 1) but inhabit overlapping climate envelopes and ecological niches in modern-day East Asia.
Despite adding three hand-picked characters (from a set of at least 25 at hand, Crepet & Nixon 1989) and accepting a phylogeny closer to the reality, the Castanopsis "clade" in the new "scaffold tree" including the Patagonian fossil remains unsupported by any exclusive or even shared and stable derived trait/set of traits (as in the original study, Wilf et al. refrain from establishing any sort of node or branch support, or test of alternative placements).
Moreover, it is safe to assume that when one adds the extinct genus Castanopsoidea to the scaffold (Wilf et al. deliberately chose not to do so), it would compete with Castanopsis rothwellii for the placement next to the modern-day Castanopsis. According to Crepet & Nixon 1989, fig. 1, one possible placement of Castanopsoidea is a sister to "Castanopsis (1)". This is not necessarily because they share a direct common origin but because these fossils also lack uniquely derived characters or a clearly derived character suite defining all Fagaceae genera except for Castanopsis (which in Crepet & Nixon's morpho-tree, is paraphyletic to Lithocarpus, which, back then, included the potential oak sister genus Notholithocarpus — literally: the 'false Lithocarpus'). Personally, for the same reasons as outlined and applied in Bomfleur et al. 2017, PeerJ 5: e3433 (and like Denk et al. 2019), I would have no problem calling all these fossils Castanopsis by defining the genus as explicitly paraphyletic, which could include the modern-day species of Castanopsis (which are probably monophyletic) and Castanopsis-like fossils that may be more or less related to them and/or other core Fagaceae: the precursors and extinct but similar, underived sister lineages.





No comments:
Post a Comment