In my previous two posts on Georges-Louis Leclerc, comte de Buffon, and his original dog genealogy of 1755, and the model for it, my interest was in Buffon's pioneering spirit in developing new ideas about genealogies and their presentation. However, it also seems natural to wonder how much we have progressed in the 250 years since then.
Having looked at the recent literature, there currently seem to be three distinct trends within dog phylogenetics:
- the study of whole-genome data, in which the results are presented solely as a neighbor-joining tree
Parker et al. (2004)
von Holdt et al. (2010) - the study of mtDNA sequence data, in which the results are presented both as a tree and as a haplotype network
Brown et al. (2011)
Kropatsch et al. (2011)
Oskarsson et al. (2012)
Ryabinina (2006) - the study of combined Y-chromosome and mtDNA sequence data, in which the results are presented solely as a haplotype network
Leonard et al. (2002)
Li et al. (2011)
Pires et al. (2006)
Savolainen et al. (2002)
Savolainen et al. (2004)
Sundqvist et al. (2006)
Verginelli et al. (2005)
I have shown the two dog trees here. Both of them make it abundantly clear, even to the casual observer, that a tree is inappropriate for the data at hand.
 |
| Dog phylogeny (Parker et al. 2004) [Click to view] |
The tree from Parker et al. has extremely small bootstrap values for almost all of the branches (only those >50% are shown on the tree), and even the group of modern dog breeds does not get up to 50% support. Clearly, there is massive conflict in this dataset. [Do not ask me why there is a value of 100% for the single branch at the base of the tree, since its presence is illogical.]
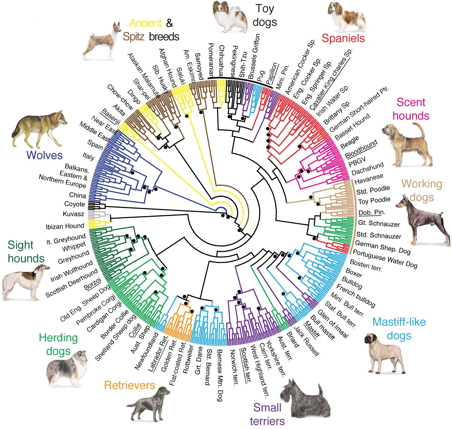 |
| Dog phylogeny (von Holdt et al. 2010) |
The tree from von Holdt et al. has broader coverage but is even more clearly non-tree-like. The dots indicate the branches with >95% bootstrap support and the colours indicate the 10 groups of dog breeds recognized by the Fédération Cynologique Internationale. As you can see, many of the breeds are scattered around the genetic tree, indicating cross-breeding in the genealogical history. This paper thus follows Buffon by nominating representative breed groups but fails by not showing the cross-breeding. So, it is drawn as a tree not a network, even when we know the history is not a tree. The use of colouring in the phylogenetic tree is one interesting way to indicate cross-connections in the genealogy, but cross-connecting lines is more explicit. [Interestingly, later editions of Buffon's work sometimes used hand-colouring of the genealogy to emphasize the breed groups that Buffon discusses in his text, so even this is not original.]
In both of these cases the tree analysis seems wildly inappropriate. As Buffon wisely told us 250 years ago, domestic dog breeds do not have a simple tree-like ancestry. It almost seems insulting that 2.5 centuries later we are still trying to fit these very same breeds (plus their numerous more-recent descendant breeds) into the straightjacket of a tree. We need to learn from the past if we are to progress into the future.
By the way, the patterns discussed here for phylogenetic analysis seem to be true for all groups of domesticated organisms. [You could try searching for the horse genealogy on the web, and you will see what I mean.] I am thus using the dogs merely as one convenient example. Following Andersen (1990), I do not intend "to pillory the few for errors which many commit with impunity".
Added note:
Since writing this post, another paper has appeared that can be added to group 1 (whole-genome data, with the results presented solely as a neighbor-joining tree): Larson et al. (2012).
References
Andersen B. (1990) Methodological Errors in Medical Research: an Incomplete Catalogue. Blackwell Science, Oxford.
Brown S.K. et al. (2011) Phylogenetic distinctiveness of Middle Eastern and Southeast Asian village dog Y chromosomes illuminates dog origins. PLoS One 6(12): e28496.
Kropatsch R. et al. (2011) On ancestors of dog breeds with focus on Weimaraner hunting dogs. Journal of Animal Breeding and Genetics 128: 64–72.
Larson G et al. (2012) Rethinking dog domestication by integrating genetics, archeology, and biogeography. Proc Natl Acad Sci USA 109: 8878-8883.
Leonard J.A. et al. (2002) Ancient DNA evidence for Old World origin of New World dogs. Science 298: 1613–1616.
Li Y. et al. (2011) The origin of the Tibetan Mastiff and species identification of Canis based on mitochondrial cytochrome c oxidase subunit I (COI) gene and COI barcoding. Animal 5: 1868-1873.
Oskarsson M.C.R. et al. (2012) Mitochondrial DNA data indicate an introduction through mainland Southeast Asia for Australian dingoes and Polynesian domestic dogs. Proceedings of the Royal Society B 279: 967-974.
Parker G. et al. (2004) Genetic structure of the purebred domestic dog. Science 304: 1160-1164.
Pires A.L. et al. (2006) Mitochondrial DNA sequence variation in Portuguese native dog breeds: diversity and phylogenetic affinities. Journal of Heredity 97: 318-330.
Ryabinina O.M. (2006) Genetic diversity and phylogenetic relationships in groups of Asian Guardian, Siberian Hunting and European Shepherd dog breeds. Proceedings of the Fifth International Conference on Bioinformatics of Genome Regulation and Structure, Volume 3, 50.
Savolainen P. et al. (2002) Genetic evidence for an East Asian origin of domestic dogs. Science 298: 1610–1613.
Savolainen P. et al. (2004) A detailed picture of the origin of the Australian dingo, obtained from the study of mitochondrial DNA. Proc Natl Acad Sci USA 101: 12387-12390.
Sundqvist A.-K. et al. (2006) Unequal contribution of sexes in the origin of dog breeds. Genetics 172: 1121–1128.
Verginelli F. et al. (2005) Mitochondrial DNA from prehistoric canids highlights relationships between dogs and south-east European wolves. Molecular Biology & Evolution 22: 2541-2551.
von Holdt B.M. et al. (2010) Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464: 898-902.

Very interesting, thanks! Phylogenetic trees may or may not be an adequate representation of evolution in some cases but it seems quite clear that they are completely inadequate to represent the divergence of "breeds" and, in particular, artificially selected breeds.
ReplyDeleteI skimmed the paper from Parker et. al and it seems that the wolf "taxa" is actually 8 different individuals. Perhaps the 100% support is simply for the split between those 8 wolves and the dog samples?
The inadequacy of a tree seems to be Buffon's main point in his genealogy. That is what makes the "first" phylogeny so interesting — we started with networks and only now are we returning to them.
DeleteIndeed, your explanation for the 100% support seems to be the correct one: "Wolves from eight different countries were combined into one population for simplicity on the tree ... When taken as individuals, all wolves split off from a single branch, which falls in the same place as the root." The countries were: China, Oman, Iran, Sweden, Italy, Mexico, Canada and the United States. Thanks for pointing that out.