Given the number of things that we can't predict in life, weather forecasting actually seems to be pretty successful, really. It's certainly better than random.
However, you rarely see any official assessments of the forecasts from any government weather bureaus. These bureaus keep records of their forecasts, and use them to refine their forecasting equations, but they rarely release any information about their perceived success rates. They do, however, release all of their data, and so we could make assessments for ourselves.
So, I thought that I might take a look at this topic for my own local area, Uppsala in Sweden. This has nothing to do with networks, which is the usual topic of this blog.
Background
"One need only think of the weather, in which case the prediction even for a few days ahead is impossible."
― Albert Einstein
The difference between
prediction and
forecasting is pretty simple. Forecasting says: "If things continue the way they have in the past, then this is what will happen next." Prediction leaves out the caveat, and simply declares: "This is what will happen next." So, technically, "weather forecasting" is not the same as "weather prediction", and the various weather bureaus around the world insist that what they are doing is forecasting
not prediction. They do not have a crystal ball, just a bunch of equations.
In some parts of the world the weather is easier to forecast than in others. In a Mediterranean-type climate, for example, we can be pretty sure that it won't rain much during summer, because that is how a Mediterranean climate is defined — hot dry summers and cool wet winters. Similarly, forecasting rain during the rainy season in the tropics is pretty straightforward. What is of more interest, then, is weather forecasting in less consistent locations.
For instance, Sydney lies at the boundary of a subtropical climate (to the north, with hot wet summers and cool dry winters) and a Mediterranean-type climate (to the south, with hot dry summers and cool wet winters). So, Sydney can have hot wet summers or hot dry summers, and cool wet winters or cool dry winters (although rarely in the same year). When there is a cool dry winter followed by a hot dry summer then Sydney makes it into the international news, due to extensive wildfires. This situation makes weather forecasting more challenging.
Oddly enough, it is quite difficult to find out just how good weather forecasting actually is, because there are not many data available, at least for most places. So, I thought I should add some.
Available Information
Most government-funded meteorological services claim to be accurate at least 2-3 days ahead, but few provide any quantitative data to back this up. There are a number of private services that provide forecasts months or even years ahead, but these provide no data at all.
The MetOffice in the U.K. claims to be "consistently one of the top two operational services in the world", and it does have a web page discussing
How accurate are our public forecasts? Their current claims are:
- 93.8% of maximum temperature forecasts are accurate to within +/- 2°C on the current day, and 90% are accurate to within +/- 2°C on the next day
- 84.3 % of minimum temperature forecasts are accurate to within +/- 2°C on the first night of the forecast period, and 79.9% are accurate to within +/- 2°C on the second night
- 73.3% of three hourly weather is correctly forecast as 'rain' on the current day, and 78.4% is correctly forecast as 'sun'.
Of perhaps more interest are independent tests of these types of claim, which are intended to compare forecasts by different providers. Unfortunately, the most ambitious of these in the U.K., the
BBC Weather Test, foundered in 2012 before it even got started, due to politics.
However, in the U.S.A. there is the
ForecastAdvisor website:
- We collect over 40,000 forecasts each day from Accuweather, CustomWeather, the National Weather Service, The Weather Channel, Weather Underground, and others for over 800 U.S. cities and 20 Canadian cities and compare them with what actually happened. All the accuracy calculations are averaged over one to three day out forecasts. The percentages you see for each weather forecaster are calculated by taking the average of four accuracy measurements. These accuracy measurements are the percentage of high temperature forecasts that are within three degrees of what actually happened [3°F = 1.7°C], the percentage of low temperature forecasts that are within three degrees of actual, the percentage correct of precipitation forecasts (both rain and snow) for the forecast icon, and the percentage correct of precipitation forecasts for the forecast text.
Thus, they present only a single "accuracy" figure for each forecaster for each location. Their example of an easy-to-forecast location (Key West, Florida) currently has a last-year average accuracy of c. 80%, while their example of a difficult one (Minot, North Dakota) has an average accuracy of 65-70%. Note that this is much lower than claimed by the U.K. MetOffice — the U.S.A. is much larger and has much more variable weather.
The ForecastAdvisor website has, however, calculated a national U.S. average for the year 2005, based on forecasts for 9-day periods (forecasts are collected at 6 pm) (
Accuracy of temperature forecasts). The average accuracy for the next-day forecast maximum temperature was 68% and the minimum temperature was 61%. (The minimum has a lower accuracy because the forecast is for 12 hours later than the forecast high.) These figures drop to 36% and 34% for the ninth-day forecast. By comparison, using the climatology forecast (ie. "taking the normal, average high and low for the day and making that your forecast") produced about 33% accuracy.
This site also has a map of the U.S.A. showing how variable were the
weather forecasts for 2004 — the more blue an area is, the less predictable weather it has, and the more red, the more predictable.
Occasionally, there are direct comparisons between the weather forecasts from different meteorological institutes. For example, the YR site of the Norwegian Meteorological Institute has been claimed to produce more accurate forecasts for several Norwegian cities than does the Swedish Meteorological and Hydrological Institute (
YR best in new weather test).
There have also occasionally been comparisons done by individuals or groups. For example, for the past 12 years the
Slimy Horror website has been testing the BBC Weather Service 5-day forecast for 10 towns in the U.K. The comparison is simplistic, based on the written description ("Partly Cloudy", "Light Rain", etc). The forecast accuracy over the past year is very high (>95%), but the long-term average is not (40-60%). The climatology forecast provided for comparison is about 35%.
Finally, in 2013, Josh Rosenberg had a look at the possibility of extending 10-day forecasts out to 25 days, and found the same as everyone else, that it is not possible in practice to forecast that far ahead (
Accuweather long-range forecast accuracy questionable).
Uppsala's Weather
Uppsala is not a bad place to assess weather forecasts. The seasons are quite distinct, but their time of arrival can be quite variable from year to year, as can their temperatures. There are rarely heavy downpours, although snowstorms can occur in winter.
Just as relevantly, Uppsala has one of the longest continuous weather records in the world, starting in 1722. The recording has been carried out by Uppsala University, and the averaged data are available from its
Institutionen för geovetenskaper. This graph shows the variation in average yearly temperature during the recordings, as calculated by the Swedish weather bureau (SMHI — Sveriges meteorologiska och hydrologiska institut) — red was an above-average year and blue below-average.
I recorded the daily maximum and minimum temperatures in my own backyard from 16 March 2013 to 15 March 2014, as well as noting the official daily rainfall from SMHI. (
Note: all temperatures in this post are in °C, while rainfall is in mm.)
Thus, recording started at what would normally be the beginning of spring, as defined meteorologically (ie. the first of seven consecutive days with an average temperature above zero). (
Note: temperature is recorded by SMHI every 15 minutes, and the daily average temperature is the mean of the 96 values each day.)
This next graph compares my maximum and minimum temperature readings with the daily average temperature averaged across the years 1981–2010 inclusive, as recorded by SMHI.
Note that there was a late start to spring in 2013 (c. 3 weeks late) and an early start to spring in 2014 (c. 4 weeks early), compared to the 30-year average. There was also a very warm spell from the middle of December to the middle of January.
Just for completeness, this next graph compares the 1981-2010 monthly data (SMHI) with the long-term data (Uppsala University). The increase in the recent temperatures is what is now called Global Warming.
Forecasting Organizations
For the primary assessment, I used two different government-funded temperature forecasts. Both of them have a forecast for the maximum and minimum temperature on the current day, plus each of the following eight days (ie. a total of nine days). I noted their forecasts at c. 8:30 each morning.
The first assessment was for the Swedish weather bureau (SMHI — Sveriges meteorologiska och hydrologiska institut). I used the forecast for
Uppsala, which is usually released at 7:45 am. SMHI provides a smoothed graphical forecast (ie. interpolated from discrete forecasts), from which the maximum and minimum can be derived each day.
The second assessment was for the Norwegian weather bureau (NMI — Norska meteorologisk institutt, whose weather site is actually called YR). I used the forecast for
Uppsala-Näs, which is usually released at 8:05 am. YR provides a smoothed graphical forecast for the forthcoming 48 hours, and a table of discrete 6-hourly forecasts thereafter.
I also used two baseline comparisons, to assess whether the weather bureaus are doing better than random forecasts. The most basic weather forecast is
Persistence: if things continue the way they are today. That is, we forecast that tomorrow's weather will be the same as today's. This takes into account seasonal weather variation, but not much else. A more sophisticated, but still automatic, forecast is
Climatology: if things continue the way they have in recent years. That is, we forecast that tomorrow's weather will be the same as the average for the same date over the past xx number of years. This takes into account within-seasonal weather variation, but not the current weather conditions. The climatology data were taken from the
TuTiempo site, averaged over the previous 12 years, with each day's temperatures being a running average of 5 days.
In addition to the SMHI and NMI forecasts, which change daily depending on the current weather conditions, I assessed two long-range forecasts. These forecasts do not change from day to day, and can be produced years in advance. In general, they are based on long-term predictable patterns, such as the relative positions of the moon, sun and other nearby planets. For example, the weather forecast for any given day might be the same as the weather observed for those previous days that the moon and sun were in the same relative positions.
The first of these long-range weather forecasts was from the
WeatherWIZ site, which claims "a record of 88 per cent accuracy since 1978", based on
this methodology. I used the forecast daily maximum and minimum temperatures for Uppsala.
The second long-range weather forecast came from the
DryDay site. This site uses an undescribed proprietary method to forecast which days will be "dry". Days are classified into three groups based on the forecast risk of rain (high, moderate, low), with "dry" days being those with a low risk that are at least one day away from a high-risk day. Forecasts are currently available only on subscription, but at the time of my study they were freely available one month in advance. I used the forecast "dry" days for Uppsala, starting on 20 May 2013 (ie. 300 days instead of the full year). For comparison, I considered a day to be non-dry if > 0.2 mm rain was recorded by SMHI in Uppsala.
It is important to note that I have not focused on rainfall forecasts. This is because rainfall is too variable locally. I well remember walking down a street when I was a teenager and it was raining on one side but not the other (have a guess which side I was on!). So, assessment of rainfall forecasting seems to me to require rainfall records averaged over a larger area than merely one meteorological station.
Temperature Forecasts
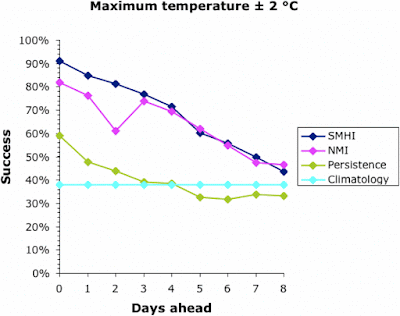
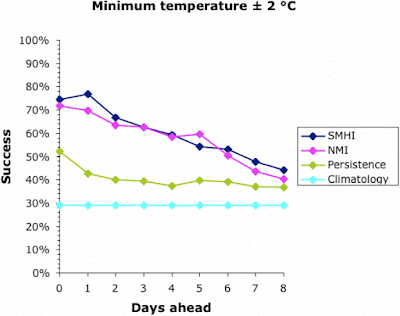
We can start to assess the data by looking at a simple measure of success — the percentage of days on which the actual temperature was within 2°C of that forecast. This is shown for all four forecasts in the next two graphs, for the maximum and minimum temperatures, respectively.
Note that the success of the baseline Climatology forecasts remained constant irrespective of how far ahead the forecast was, because it is based on previous years' patterns not the current weather. The success of the other forecasts decreased into the future, meaning that it is easier to forecast tomorrow than next week. All forecasts converged at 30-40% success at about 9 days ahead. This is why most meteorological bureaus only issue 10-day forecasts (including the current day). This, then, defines the limits of the current climatology models for Uppsala; and it matches those quoted above for the U.K. and U.S.A.
Interestingly, the success of all forecasts was better for the maximum temperature than the minimum, except for the Persistence baseline which was generally the other way around. This remains unexplained. The Persistence baseline was generally a better forecaster than the Climatology one; after all, it is based on current weather not previous years'. However, for the maximum temperature this was only true for a couple of days into the future.
Both of the meteorological bureaus did consistently better than the two baseline forecasts, although this decreased consistently into the future. Sadly, even forecasting the current day's maximum temperature was successful to within 2°C only 90% of the time, and the minimum was successful only 75% of the time. This also matches the data quoted above for the U.K. and U.S.A.
Both bureaus produced better forecasts for the maximum temperature than for the minimum. The SMHI forecast was better than the NMI for the first 2–3 days ahead, but not after that. The dip in the NMI success occurred when changing from the smoothed hourly forecasts to the 6-hour forecasts, which suggests a problem in the algorithm used to produce the web page.
We can now move on to considering the actual temperature forecasts. The next two graphs show the difference between the actual temperature and the forecast one, averaged across the whole year. For a perfect set of forecasts, this difference would be zero.
The Climatology baseline forecasts overestimated both the maximum and minimum temperatures, which suggests that the recording year was generally colder than average. Some replication of years is obviously needed in this assessment. The Persistence baseline increasingly underestimated the future temperature slightly. This implies that the future was generally warmer than the present, which should not be true across a whole year — perhaps it is related to the presence of two unusually warm spells in 2014.
Both bureaus consistently under-estimated the maximum temperature and over-estimated the minimum. NMI consistently produced lower forecasts than did SMHI. Thus, NMI did better at forecasting the minimum temperature but worse at forecasting the maximum. Interestingly, the difference between the forecast and actual temperature did not always get worse with increasing time ahead.
Finally, we should look at the variability of the forecasts. The next two graphs show how variable were the differences between the actual temperature and the forecast one, taken across the whole year.
Other than for Climatology, the forecasts became more variable the further they were into the future. There was no difference between the two bureaus; and, as noted above, their forecasts converged to the Climatology baseline at about 9 days ahead. The Persistence baseline forecasts were usually more variable than this.
Overall, the meteorological bureaus did better than the automated forecasts from the baseline methods. That is, they do better than merely forecasting the weather based on either today or recent years. However, there were consistent differences between the actual and forecast temperatures, and also between the two bureaus. Their models are obviously different; and neither of them achieved better than a 75-90% success rate even for the current day.
Long-term Forecasts
This next graph shows the frequency histogram of the long-range temperature forecasts from the WeatherWIZ site, based on 5-degree intervals (ie. 0 means –2.5 < °C < +2.5).
The forecasts were within 5°C of the actual temperature 68% of the time for the maximum and 62% for the minimum, with a slight bias towards under-estimates. This bias presumably reflects the higher temperatures in recent years, compared to the data from which the forecasts were made. (Has anyone commented on this, that long-range forecasts will be less accurate in the face of Global Warming?)
The WeatherWIZ forecasting result seems to be remarkably good, given that the current weather is not taken into account in the forecast, only long-term patterns. This does imply that two-thirds of our ability to forecast tomorrow's weather has nothing to do with today's weather, only today's date.
However, the forecasts were occasionally more than 15°C wrong (–13.2 to +16.2 for the maximum temperature, and –14.2 to +18.8 for the minimum). This occurred when unseasonable weather happened, such as during the mid-winter warm spell. So, the one-third of forecast unpredictability can be really, really bad — today's weather is not irrelevant!
The rainfall forecasts, on the other hand, were not all that impressive (based on the 300 days rather than the whole year). This is not unexpected, given the locally variable nature of rain.
If we classify the DryDay forecasts as true or false positives, and true or false negatives, then we can calculate a set of standard characteristics to describe the "dry" day forecasting success:
Sensitivity (true positive rate) =
Specificity (true negative rate) =
Precision (positive predictive value) =
Accuracy =
|
33.8% actual dry days were correctly forecast
81.8% actual non-dry days were correctly forecast
67.1% forecasts were correct
56.7% forecast "dry" days were correct |
This shows that the forecasting method actually does better at predicting non-dry days than dry days (61% of the days actually had <0.2 mm of rain).
However, overall, the method does better than random chance, with a Relative Risk of 0.622 (95% CI: 0.443–0.872) — that is, the chance of rain on a forecast "dry" day was 62% of that on the other days. The following ROC curve illustrates the good and the bad, with a rapid rise in sensitivity without loss of specificity (as desired), but the forecasts then become rapidly non-specific.
Conclusion
"But who wants to be foretold the weather? It is bad enough when it comes, without our having the misery of knowing about it beforehand."
― Jerome K. Jerome,
Three Men in a Boat