One may expect that perfectly compatible, trivial data will lead to perfect trees that are trivial to interpret. And this may really be the case when phylogenetics is restricted to contemporary taxa and molecular data. Adding to various earlier posts that deal with data patterns and their representation in inference graphs (e.g. Networks can outperform PCA..., Stacking neighbour-nets..., Clades, cladograms, cladistics ... and networks ...), I will show in this post what we get when we deal with very trivial, straightforward to interpret, data.
Two trivial scenarios: a linear and a dichotomous evolutionary sequence
The virtual data matrix for our experiment comprises seven taxa (OTUs) from different time scales and six binary (Dollo) characters. There are two historical scenarios that are supported by patterns in the data (see the first figure).
The linear scenario has a mother taxon that evolves by acquiring a unique, persistent trait, and is replaced by its daughter taxon through time. In contrast, the dichotomous scenario has two subsequent events of cladogenesis: the all-ancestor A splits into two taxa (B, E), each defined by a unique change in a binary character passed on to their descendants. B and E then underwent a second cladogenetic event, giving rise to C+D and F+G.
The resultant data matrices have different properties. In the case of the linear evolution, all changes lead to synapomorphies sensu Hennig (characters #1–#5) along with one terminal autapomorphy of the latest member of the lineage, G (character #6).
In the case of the dichotomous evolution, we have two synapomorphies supporting the BCD and EFG clades (characters #1, #4), respectively, and four autapomorphies (each one for C, D, F and G, the youngest set of taxa).
The following figure shows the character-based splits (taxon bipartitions) for the linear evolution scenario:
 |
| (Trivial splits, one taxon separated from all others, in blue) |
Reconstructing the (true) evolutionary pathway is trivial based on this perfect split pattern, especially if we know that A is the oldest taxon and G the youngest.
It's equally straighforward for our second scenario, with perfectly dichotomous evolution:
 |
Character 1 and character 4 define taxon cliques comprising B,C,D and E,F,G. The remaining characters indicate that C,D and F,G derive from B and E, respectively. |
 |
Explicit inferences
As stated above, the data properties for both scenarios are different. The matrices have a different number of parsimony-informative characters (4 for linear, 2 for dichotomous). Accordingly, the reconstructed optimal trees (here using the maximum parsimony, least-squares, and maximum likelihood criteria), are better resolved / more correct for the linear than for the dichotomous evolution.
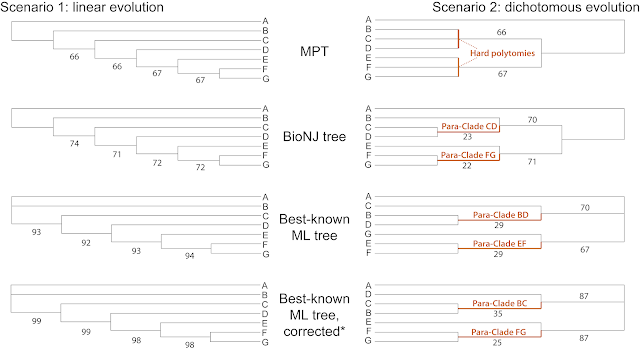 |
| MPT = most-parsimonious tree; ML = maximum likelihood. *Corrected for ascertainment bias. |
Using all of the variable characters, NJ and ML are generally more decisive and produce higher support for the right branches. But for the dichotomous evolution scenario, they also show ghost-clades ("para-clades" as they include close relatives sharing a recent common origin, but do not represent monophyletic groups sensu Hennig) with low support. The corresponding MPT has no ghost-clades, but it also provides no clues to how B,C,D and E,F,G are related to each other.
Beyond this, and as can be seen in many real-world examples, there is no fundamental difference between character-based inferences such as maximum parsimony (MP) or maximum likelihood (ML) and distance-based inferences (NJ) fulfilling (here) the least-squares criterion (sometimes still called "phenetic" inferences in contrast to the "phylogenetic" parsimony, Bayesian inference and maximum likelihood).
The differences diminish further when we look at the phylograms instead of the cladograms, as shown next.
Another observation we can make is that for the linear-evolution scenario (four synapormophies), the ascertainment bias correction under ML has little effect, but it is crucial for the dichotomous evolution (two synapomorphies) to get sensible branch lengths.
Parsimony provides the most conservative (and least decisive) results for the dichotomous-evolution scenario, also because of the way I applied it: PAUP* allows optimizing trees with hard polytomies when using the default branch-and-bound search (for tree inference as well as bootstrapping), whereas the NJ / BioNJ algorithm and the ML implementation in RAxML will always produce fully dichotomized trees, including zero-length or near-zero-length branches. This explains the difference in the support values of preferred and alternative splits.
 |
(Non-filtered) Bootstrap support consensus networks for the linear evolution scenario. Same scale for all graphs, trivial splits (dashed lines) collapsed. |
 |
| (Non-filtered) Bootstrap support consensus networks for the dichotomous evolution scenario. |
Trees are not wrong, but they miss the point
None of the graphs above show anything strongly erroneous, but they also don't fully capture the evolutionary pathways — that is, the actual ancestor-descendant relationships. This is because our taxon set includes ancestral forms, which, in traditional trees, have to be placed as sisters to part or all of their descendants. Networks provide a quick solution to this limitation.
 |
| Median-joining networks inferred with NETWORK 5.0.0.3 for both scenarios, with the inferred (and real) character changes annotated along edges. |
 |
| Neighbour-nets inferred with SplitsTree 4.13.1 for both scenarios, based on the mean (Hamming) pairwise distances. |
The two (perfectly tree-like) graphs, one parsimony-based, the other distance-based, look identical, and place all of the taxa exactly where they should be: the ancestors on the nodes ("medians"), and their (latest) descendants at the tips. But note that in the case of the Neighbour-net this is a visual illusion / approximation: in fact, the ancestors are actually connected by zero-length edges to the node they appear to be sitting on.
Given that both scenarios used here produce trivial, straightforward to interpret, data patterns (see the first figures), the failure of the traditional tree inferences to get it completely right can be a bit unsettling. Trees including primitive-old and derived-new forms are common in the (palaeontological) literature, and typically show many branches lacking high support (note that only ML produced a bootstrap support >90 for a true-tree branch, and only for the linear evolution scenario). To address evolution over time, networks should hence be standard applications, rather than the exception. Cladograms should be long gone, as they show very little beyond the most trivial.
If we want trees (and many of us want trees!), we need tree inferences that can optimize an older taxon on an internal branch or node, to accommodate potentially ancestral forms.
Related blog posts
In Clades, cladograms, cladistics, and why networks are inevitable, I argue that we cannot get around networks when we aim to study taxa from different time scales using their morphologies.
Digging deeper: Population dynamics and individual-based fossil phylogenies raises the question of what we deal with when we use individual fossils (i.e. long-dead individuals) as OTUs in our phylogenetic inferences.
Monophyletic groups in networks by David gives an introduction into (fringe) terminology. What to do when dealing with more than a single most-recent common ancestor and past reticulation?
Networks and most recent common ancestors by David discusses the concepts of conservative MRCAs (most recent common ancestors), fuzzy MRCAs and (alternative) LCA — lowest (last) common ancestors in the face of reticulation.
In Stacking neighbour-nets: ancestors and descendants, I outline how one may (and why one should) stack Neighbour-nets to analyse the evolutionary history of a group including (mostly) fossil representatives.
The first Darwinian evolutionary tree[s] show features one rarely finds in a modern-day phylogenetic tree: ancestral and descendant forms, ancestral taxa addressed as species and not higher taxa, and gradual transition between forms (post by David).
Tree metaphors and mathematical trees by David, which introduces János Podani's concept about "branching silhouettes" and how to depict an actual evolutionary tree.
Where have all the ancestors gone? discusses the common notion that we don't have to deal with ancestor-descendant problems in phylogenetics at all, because the scarcity of the (terrestrial) fossil records ensures to only find extinct side (sister) lineages.




For those readers not familiar with networks, I proposed a parsimony-based tree-based method for infering ancestor-descendant relationships : http://ukrbotj.co.ua/archive/74/2/103
ReplyDeleteAs you said, "we need tree inferences that can optimize an older taxon on an internal branch or node, to accommodate potentially ancestral forms", this is exactly what my algorithm is doing.
Dear Damian,
ReplyDeleteI still like the "commagram" (even though it's a phylogenetic tree, there are no reticulations), but the main problem (I can't judge the maths) with your method is
a) that it's published in the Ukrainian Botanical Journal (so I suppose the number of people picking it up will be quite limited), and
b) that it doesn't come with a programme/script (as far as I can see) doing the Bayesian weighting and inferring the so-optimised tree that includes likely and theoretical ancestors.
In order to have people play around with it, you could upload the NEXUS files you used for PAUP*, a walk-through, as well as the graphics on an open repository, e.g. figshare or maybe, in this particular case, PaleorXiv. You have a similar problem here as Zander had/has (I always liked his papers, like this one) with his ideas. Providing a theoretical-philosophical piece for a practical thing that could interest mainly practioners with no/little mathematical background whatsoever: palaeontologists.
This is why everyone uses the post-analysis weighting method implemented in the WHS-supported TNT to get some resolved tree in palaeonotlogy, despite many ancestor-descendant and other signal issues in their data. Because it's just pressing buttons these days.
Cheers, Guido