Wednesday, May 28, 2014
Phylogenetic networks and "evolutionary networks"
Complex networks are found in all parts of biology, graphically representing biological patterns and, if they are directed networks, also their causal processes. Directed networks are currently used to model various aspects of biological systems, such as gene regulation, protein interactions, metabolic pathways, ecological interactions, and evolutionary histories.
Two types of networks can be distinguished, and this distinction seems to me to be very important. Most networks are what might be called observed networks, in the sense that the nodes and edges represent empirical observations. For example, a food web consists of nodes representing animals with connecting edges representing who eats whom. Similarly, in a gene regulation network the genes (nodes) are connected by edges showing which genes affect the functioning of which other genes. In all cases, the presence of the nodes and edges in the graph is based on experimental data. These are collectively called interaction networks or regulation networks.
However, when studying historical patterns and processes not all of the nodes and edges can be observed. So, instead, they are inferred as part of the data-analysis procedure. That is, we infer the patterns as well as the processes; and we can call these inferred networks. In this case, the empirical data may consist solely of the leaf nodes, and we infer the other nodes plus all of the edges. For example, every person has two parents, and even if we do not observe those parents we can infer their existence with confidence, as we also can for the grandparents, and so on back through time with a continuous series of ancestors. Alternatively, we may also observe some of the internal nodes of the network, such as when we do record the parents and grandparents because they are contemporaneous (ie. their generations overlap). This type of pattern can be represented as a genealogical network, when referring to individual organisms, or a phylogenetic network when referring to groups (populations, species, or larger taxonomic groups).
What, then, are the things often referred to as "evolutionary networks" but which are clearly not phylogenetic networks? They are of the first type, the interaction networks. In an evolutionary network the observed nodes are directly connected to each other to represent some aspect of evolution. This aspect may have some component of phylogeny to it, but there is more to the study of evolution than solely phylogenetic history.
For example, directed LGT (dLGT) networks connect nodes representing contemporary organisms with edges that represent inferred lateral gene transfer. That is, the evolutionary networks show gene sharing. This is obviously related to the phylogeny of the organisms, but the network does not display the phylogeny itself. This first example (from Ovidiu Popa, Einat Hazkani-Covo, Giddy Landan, William Martin, Tal Dagan. 2011. Directed networks reveal genomic barriers and DNA repair bypasses to lateral gene transfer among prokaryotes. Genome Research 21: 599-609) shows "32,028 polarized lateral recipient–donor protein-coding gene transfer events" inferred from "the completely sequenced genomes of 657 prokaryote species".
The concept of a gene-sharing network as an evolutionary network has also been applied to viruses and their relatives, for example, as shown by this next diagram (from Natalya Yutin, Didier Raoult, Eugene V Koonin. 2013. Virophages, polintons, and transpovirons: a complex evolutionary network of diverse selfish genetic elements with different reproduction strategies. Virology Journal 10: 158).
The question, then, is what to make of diagrams that combine both a phylogenetic tree and this type of evolutionary network, such as is done in the Minimal Lateral Network. This next example is from linguistics rather than biology (from Johann-Mattis List, Shijulal Nelson-Sathi, Hans Geisler, William Martin. 2013. Networks of lexical borrowing and lateral gene transfer in language and genome evolution. Bioessays 36: 141-150), and it superimposes the sharing network and the phylogenetic tree. (For a discussion in the context of LGT, see also Tal Dagan. 2011. Phylogenomic networks. Trends in Microbiology 19: 483-491).
In this diagram, the tree explicitly represents the phylogenetic history of the languages while the evolutionary network represents possible borrowings of words, with thicker lines representing more borrowed words. Clearly, the network also contains phylogenetic information of some sort. For example, the connection of the root of the Romance languages to English reflects the conquest of Britain by the French-speaking Normans, which modified the Old-German heritage of Old English. However, the diagram as a whole is a hybrid, rather than being a coherent phylogenetic network in the simplest sense (ie. a reticulation network).
To see this clearly, note that the phylogenetic tree is not fully resolved and that the evolutionary network does suggest possible resolutions for several of polychotomies, such as the relationship of Armenian and Greek, the relationship of Albanian to the Romance languages, and the relationship of the Gaelic languages to the Romance languages. So, in some cases the evolutionary network helps resolve the phylogenetic tree rather than forming a reticulating network.
It would be possible to derive a phylogenetic network from this minimal lateral network, but as it stands it is a combination of a phylogenetic tree and a so-called evolutionary network.
Labels:
DAG,
Evolutionary network,
Phylogeny
Monday, May 26, 2014
Tattoo Monday IX
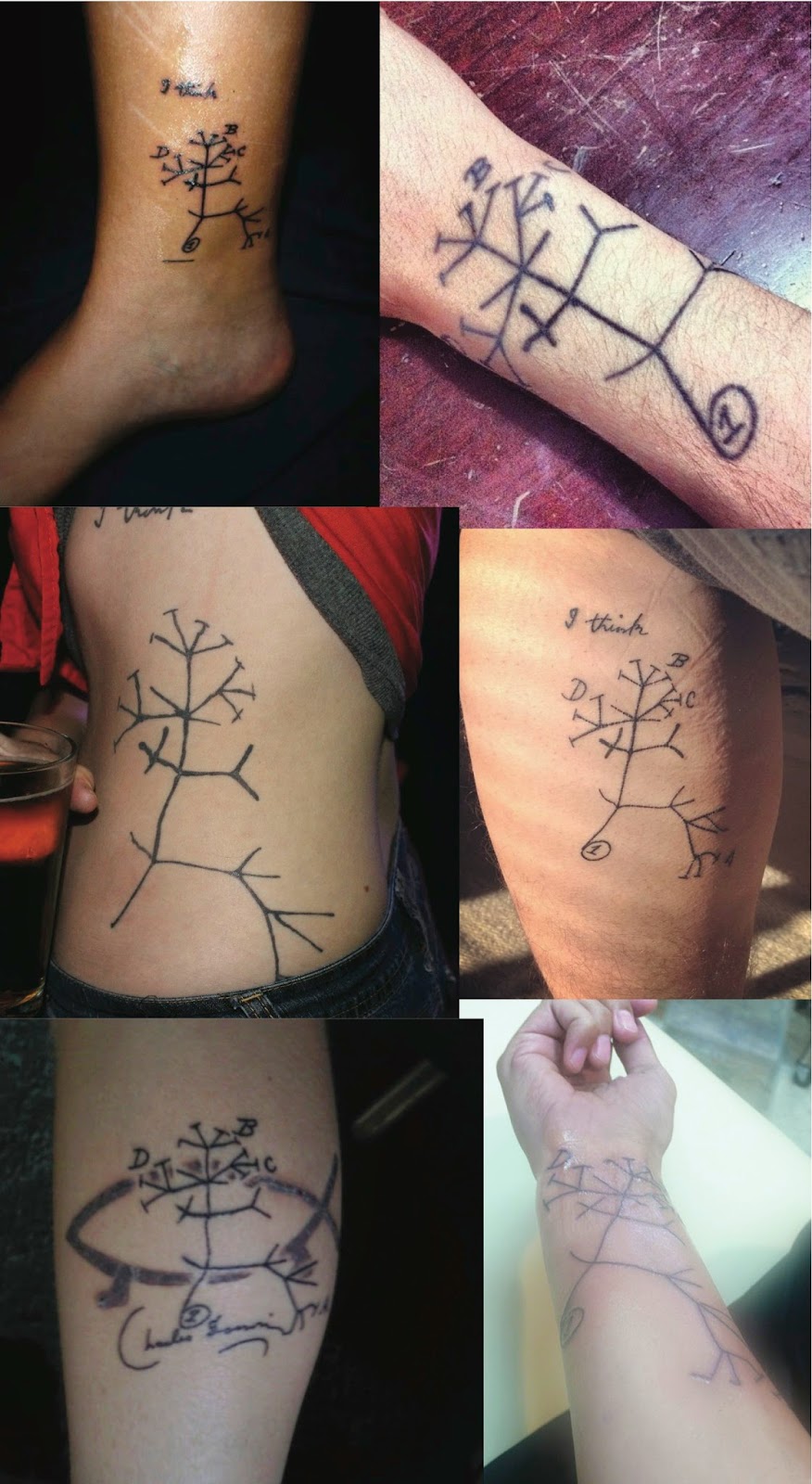
We haven't had any phylogenetic tree tattoos on this blog for a while, so here is a new collection of Charles Darwin's best-known sketch from his Notebooks (the "I think" tree) (for other examples, see Tattoo Monday III, Tattoo Monday V, and Tattoo Monday VI).
Labels:
Tattoo
Wednesday, May 21, 2014
Phylogenetics of computer viruses?
There is a difference between phylogenetics and clustering or classification. The latter processes put objects into groups based on some intrinsic features, but the former uses their intrinsic features to expresses their evolutionary history. Not all objects have an evolutionary history, even though they can all be put into groups. Furthermore, even objects that do have a history do not necessarily have an evolutionary history. Evolution involves ancestor-descendant relationships (as well as sister-group relationships), and not all of history involves ancestors and descendants.
This distinction is important for the use of phylogenetics as a metaphor for the history of non-biological objects. Outside of biology, many things are claimed to have a "phylogenetic history", including languages and most human artifacts. As I have noted before, one has to be careful when applying this metaphor (see False analogies between anthropology and biology).
One particular example that I have encountered involves the development of computer viruses and other malware (Iliopoulos et al. 2008). Metaphorically, such viruses can be seen to be phylogenetically related, because new viruses are often based on previous ones — that is, one virus "begets" another virus due to changes in its intrinsic attributes. In this sense the metaphor is helpful, although there is no actual copying of anything resembling a genome — this is phenotype evolution not genotype evolution.
Sorkin (1994) seems to have been the first to discuss the possibility of computer virus evolution, but the first empirical attempt to reconstruct a digital phylogeny appears to have been by Hull (1995b), who studied the Stoned computer virus (a virus that infected the boot sector of PCs between 1990 and 1995), as shown below.
Since then, phylogenetics has been a popular topic in the study of computer malware (eg. Goldberg et al. 1996; Carrera & Erdélyi 2004; Karim et al. 2005a,b; Ma et al. 2006; Wehner 2007; Walenstein et al. 2007; Wagener et al. 2008: Hayes et al. 2009; Khoo & Lió 2011; Guan et al. 2012). As noted by Webster & Malcolm (2007) these studies "present classifications of malware based on phylogenetic trees, in which the lineage of computer viruses can be traced and a 'family tree' of viruses constructed based on similar behaviors." (Note that there are other possible uses of phylogenetics related to computer programming, as exemplified by Ji et al. 2008.)
In all cases the phylogeny was produced using a distance-based clustering algorithm (ie. the tree is a phenetic one). Some of the distances are well motivated in terms of historical changes in the intrinsic attributes of the malware, but that does not necessarily make the resulting phenogram a phylogeny. So, the methods use basic clustering techniques to produce a tree, thus treating classification as phylogenetics (or phenetics as phylogenetics). This certainly clusters the objects, but there is no necessary reason for the clustering to reflect phylogeny.
Thus, a simple concept of clustering is inadequate, even though it can be used to construct a tree. A phylogenetic tree expresses the nested hierarchy formed by the shared derived character states, but not by anything else. A tree expresses nested clusters, but it is a form of "special nesting" that expresses a phylogeny. Only one form of tree is relevant to phylogenetics, and trees formed in other ways are likely to be suitable only under the simplest circumstances.
Furthermore, the general conception in these papers of a virus phylogeny as tree-like is clear. As noted by Hull (1995a):
Computer viruses evolve in complex ways not usually encountered in nature. The transplantation of large segments of computer code from one virus to another need not represent evolutionary relationship, for example. A newer virus may just represent a debugged or patched earlier version. The virus author may have deliberately incorporated parts of other viruses as a short cut, or because the plagiarized code is useful. If the virus incorporates code generating 'engines', similar code may appear in viruses with no other similarities. Structural similarities deriving from functional similarities likewise derive from several sources.As we now know, these sorts of evolutionary events are usually found in nature, but they create reticulate histories, involving horizontal as well as vertical evolution. That is, computer virus phylogenetics involves reticulation — new computer code takes bits and pieces from various previous viruses. In particular, there is also what is called "oblique" evolution, in which there is horizontal evolution between generations. This is a characteristic of many histories involving human artifacts (see Time inconsistency in evolutionary networks), and it allows information to "time travel", so that the information available for horizontal transmission can come from the distant past as well as from the present.
So, malware evolution is not tree-like. Only two of the papers cited above seem to acknowledge this fact. Khoo & Lió (2011) were quite conventional in using splits graphs rather than unrooted trees to display their data, although they do not specify the algorithm for producing the networks. They do, however, claim that "networks were more useful for visualising short nop-equivalent code metamorphism than trees".
Goldberg et al. (1996) were more innovative, and analyzed their data using what they called a "phyloDAG", which is a directed network that can have multiple roots (it appears to be a type of minimum-spanning network). Interestingly, they note that "Beyond the computer virus realm for which it was conceived, the phyloDAG is also a plausible model for evolution of bacterial populations." Indeed, the possibility of multiple roots has been explicitly suggested for prokaryote phylogenetics (see Can networks have multiple roots?). I wouldn't doubt that it is also feasible for language history.
References
Carrera E, Erdélyi G (2004) Digital genome mapping – advanced binary malware analysis. Virus Bulletin Conference 2004.
Goldberg LA, Goldberg PW, Phillips CA, Sorkin GB (1996) Constructing computer virus phylogenies. Lecture Notes in Computer Science 1075: 253-270. [also Journal of Algorithms (1998) 26: 188-208]
Guan Q, Tang Y, Liu X (2012) A malware homologous analysis method based on sequence of system function. Advanced Science and Technology Letters, ASTL 15: Advanced Computer Science and Technology. Science and Engineering Research Support Society, Sandy Bay, Tasmania, Australia.
Hayes M, Walenstein A, Lakhotia A (2009) Evaluation of malware phylogeny modelling systems using automated variant generation. Journal in Computer Virology 5: 335-343.
Hull DB (1995a) Computer viruses: naming and classification. Virus Bulletin Sept: 15-17.
Hull DB (1995b) Computer viruses: naming and classification, part II. Virus Bulletin Oct: 16-17.
Iliopoulos D, Adami C, Ször P (2008) Darwin inside the machines: malware evolution and the consequences for computer security. Virus Bulletin Conference 2008.
Ji J-H, Park S-H, Woo G, Cho H-G (2008) Generating pylogenetic tree of homogeneous source code in a plagiarism detection system. International Journal of Control, Automation, and Systems 6: 809-817.
Karim ME, Walenstein A, Lakhotia A (2005a) Malware phylogeny using maximal pi-patterns. Proceedings of the EICAR 2005 Conference, pp 156-174.
Karim ME, Walenstein A, Lakhotia A, Parida L (2005b) Malware phylogeny generation using permutations of code. Journal in Computer Virology 1: 13-23.
Khoo WM, Lió P (2011) Unity in diversity: phylogenetic-inspired techniques for reverse engineering and detection of malware families. Proceedings of the 2011 First Systems Security Workshop (SysSec'11), pp 3-10. IEEE Computer Society Washington, DC.
Ma J, Dunagan J, Wang HJ, Savage S, Voelker GM (2006) Finding diversity in remote code injection exploits. Proceedings of the 6th ACM SIGCOMM Conference on Internet Measurement, pp 53-64. ACM, New York.
Sorkin GB (1994) Grouping related computer viruses into families. Proceedings of the IBM Security ITS 1994.
Wagener G, State R, Dulaunoy A (2008) Malware behaviour analysis. Journal in Computer Virology 4: 279–287.
Walenstein A, Hayes M, Lakhotia A (2007) Phylogenetic comparisons of malware. Virus Bulletin Conference 2007.
Webster M, Malcolm G (2007) Classification of computer viruses using the theory of affordances. Second International Workshop on the Theory of Computer Viruses.
Wehner S (2007) Analyzing worms and network traffic using compression. Journal of Computer Security 15: 303-320. (arXiv:cs/0504045v1, 2007)
Labels:
Classification,
Phylogeny,
Rooting,
Time consistency,
Viruses
Monday, May 19, 2014
Cleopatra, ambition and family networks
There is an old saying in English that "Behind every great man there is a woman ... telling him to be great". This is intended to indicate that even in patrilineal societies women have influenced history, even if history has chosen not to formally recognize them (or historians have, anyway). However, every so often a woman has also stepped into the spotlight for herself, and recognizably influenced events in a way that has brought her name down through history.
The most famous of these is probably Cleopatra (or more properly Kleopatra), the last ruler of Ancient Egypt (as Cleopatra VII). Sadly, her ambition to become Empress of the known world seems to have destroyed two successive Roman rulers (Julius Caesar and Marc Antony) as well as her own two brothers (who would have ruled in her place); and her failure seems to have lost the country of which she was queen, so that Egypt became a Roman dependency. She ruled from 51-30 BCE, and modern Egypt did not regain its independence until 1953. This was one seriously influential woman.
As noted by Schiff (2010):
She lost her kingdom once; regained it; nearly lost it again; amassed an empire; lost it all. At the height of her power she controlled virtually the entire eastern Mediterranean coast, the last great kingdom of any Egyptian ruler. For a fleeting moment she held the fate of the Western world in her hands ... Catastrophe reliably cements a reputation, and Cleopatra's end was sudden and sensational.Her interest to us, however, is her role in a dynasty that favored incest, and thus had a "family tree" that was a hybridization network, as shown in the figure. This particular family history is rather complex. Note that Cleopatra herself had at least four liaisons, two with her brothers (who successively ruled jointly with her as Ptolemy XIII and Ptolemy XIV, respectively) and two with Romans (Julius Caesar and Marc Anthony). Later, she also ruled jointly with her son by Julius Caesar (as Ptolemy XV).
 |
Adapted from the Too Much Information blog, based on the information at Ian Mladjov's Genealogical Tables |
The Ptolemaic dynasty was founded after the death of Alexander the Great (aka Alexander III of Macedon), when his empire was divided up among his Greek generals, and in 323 BCE Egypt ended up in the hands of Ptolemy, who subsequently ruled as the pharaoh Ptolemy I from 305-282 BCE. As Dray (2012) has noted:
His daughter, Arsinoe II, would start the tradition of incest. Married off to an old King of Thrace when she was still a teenager, she was the ultimate survivor. Her life was frequently in danger and she made many narrow escapes ... At some point, Arsinoe seems to have decided that if she wanted to be safe, she couldn’t trust anyone outside her immediate family. So, she returned to Egypt and married her full brother, Ptolemy II.
Now, the Greeks didn’t have a tradition of incest in their ruling families … but the pharaohs of Egypt did. By marrying her brother, Arsinoe was able to help create a link between the new Ptolemaic dynasty and the very old traditions of the native Egyptians. It served her extremely well as she became the first female pharaoh of the Ptolemaic dynasty, ruling not just as the wife of the king, but as a king in her own right.Meeg (2009) suggests that:
According to tradition, incestuous marriages between the pharaohs and their sisters were common. If this was the case, it could have been done to emulate the god Osiris and his sister / wife the goddess Isis (the product of that union was Horus, the alleged ancestor of the Pharaoh), and/or to keep the sacred bloodline pure. When Alexander the Great's general Ptolemy seized control of Egypt around 323 BC, his descendants would continue the local custom of pharaonic brother-sister marriages. This practice was unknown among Greeks and Macedonians.Indeed, Wikipedia notes:
In ancient Egypt, royal women carried the bloodlines and so it was advantageous for a pharaoh to marry his sister or half-sister; in such cases a special combination between endogamy and polygamy is found. Normally the old ruler's eldest son and daughter (who could be either siblings or half-siblings) became the new rulers. All rulers of the Ptolemaic dynasty from Ptolemy II were married to their brothers and sisters, so as to keep the Ptolemaic blood "pure" and to strengthen the line of succession. Cleopatra VII (also called Cleopatra VI) and Ptolemy XIII, who married and became co-rulers of ancient Egypt following their father's death, are the most widely known example.Bevan (1927) continues the story [Note: he uses one number less for the Cleopatras and Ptolemies]:
Cleopatra VI found herself queen of Egypt at the age of seventeen or eighteen. By the custom of the house, and according to the will and testament of Ptolemy Auletes, the elder of her two brothers, then only nine or ten, was associated with her, as king (Ptolemy XII). They probably had, as a pair, the style of "Father-loving Gods" (Theoi Philopatores), though neither during the reign of Cleopatra with Ptolemy XII, nor during her reign, later on, with the younger brother, Ptolemy XIII (then about twelve), do the coins bear any head or name but that of the queen, and in Egyptian sepulchral inscriptions put up during the reign of Cleopatra with her younger brother (regnal years 5, 6, and 7 of Cleopatra) the regnal year of the boy-king is ignored. Ptolemy XIV was the acknowledged son of Julius Caesar and Cleopatra, and ruled as child king with his mother.The involvement of royalty in consanguinity and incest is widespread. As noted by Dobbs (2010):
While virtually every culture in recorded history has held sibling or parent-child couplings taboo, royalty have been exempted in many societies, including ancient Egypt, Inca Peru, and, at times, Central Africa, Mexico, and Thailand [and also Hawaii].I have already discussed incest in the family "trees" of the Egyptian 18th Dynasty, in Tutankhamun and extreme consanguinity (the other set of pharaohs where this was common); and I have covered the persistent inbreeding in the downfall of the modern Spanish branch of the Habsburgs, in Family trees, pedigrees and hybridization networks.
Not unexpectedly, this phenomenon has received attention from modern evolutionary biologists. Conventionally, the evolutionary advantage of sexual over non-sexual reproduction is considered to be the creation of genetic diversity through heterozygosity. Inbreeding, by reducing heterozygosity, then seems to negate the advantages of sexual reproduction. So, the near universality of incest avoidance in humans has a clear genetic dimension. Indeed, as I have noted in previous blog posts this is easily demonstrated in well-known families — (i) Charles Darwin's family pedigree network, (ii) Toulouse-Lautrec: family trees and networks.
The presence of incest among royal families then requires biological explanation. Indeed, van den Berghe & Mesher (1980) have provided one:
Royal incest (mostly brother-sister; less commonly father-daughter) represents the logical extreme of hypergyny. Women in stratified societies maximize [evolutionary] fitness by marrying up; the higher the status of a woman, the narrower her range of prospective husbands. This leads to a direct association between high status and inbreeding. Royal incest is a fitness maximizing strategy if the following conditions are met: polygyny, patrilineal succession, and parental control of royal succession. Under those conditions, the genetic risks of close inbreeding are more than accounted for by the production of a highly related male heir who has, himself, access to a large harem. Data from Ancient Egypt, Inca Peru, Hawaii, Thailand, Monomotapa, Bunyoro, Ankole, Buganda, Shilluk, Zande, Nyanga and Dahomey confirm hypotheses derived from the sociobiological paradigm of inclusive fitness.Finally, to return to Cleopatra, she is usually credited with being fatally attractive due to her great beauty. However, there is no evidence that this was actually the case. Her attractiveness to men seems to have come much more from a strong personality, including determined diplomacy and an easy facility with languages. Also, her ancestors were Macedonian Greeks, rather than native Egyptians, giving her a stronger genetic and cultural tie to Europe rather than to Africa, which must have helped when trying to woo the rulers of the Roman Empire. It was this ancestry that the dynasty's consanguinity and incest were intended to protect. The Egyptian populace certainly didn't benefit from it.
Indeed, Cleopatra seems simply to have been the ultimate expression of her dynasty's heritage, as noted by Ager (2006):
royal incest, as practised by the Ptolemies, was only one of a larger set of behaviours, all of which were symbolic of power, and all of which were characterized by lavishness, immoderation, excess and the breaching of limits in general.Interestingly, the potentially negative aspects of inbreeding seem not to have affected this dynasty — there is no convincing evidence of infertility, infant mortality or genetic defects, for example (Ager 2006). Instead, their main historical legacy has been their bizarre juxtaposition of either marrying each other or murdering each other, and sometimes both. Cleopatra's activities in this regard were no different to those of her ancestors.
References
Ager SL (2006) The power of excess: royal incest and the Ptolemaic dynasty. Anthropologica 48: 165-186.
Bevan ER (1927) The House of Ptolemy. Methuen Publishing, London.
Dobbs D (2010) The risks and rewards of royal incest. National Geographic Magazine.
Dray S (2012) Keeping it in the (Ptolemaic) family: when incest is best.
Meeg (2009) Royal inbreeding in Ancient Egypt.
Ian Mladjov's Genealogical Tables — The Ptolemies, kings of Egypt.
Schiff S (2010) Cleopatra: a biography. Little, Brown and Co, New York. [excerpted in Smithsonian Magazine]
van den Berghe PL, Mesher GM (1980) Royal incest and inclusive fitness. American Ethnologist 7: 300-317.
Wikipedia. Inbreeding.
Labels:
Genealogy,
Hybridization network,
Pedigree
Wednesday, May 14, 2014
Non-model distances in phylogenetics
Bioinformatics is sometimes divorced from biology. This happens when the ideas involved are solely computational ones, and are not biologically motivated. For example, Otu et al. (2003), when proposing "a new sequence distance measure for phylogenetic tree construction" point out that "it is worth noting that our distance measures do not use any evolutionary model". They seem to see this a a Good Thing whereas biologists might not.
Pairwise distances are actually a good case in point, because there are many ways to measure the similarity / distance between any two objects, and most of them have nothing whatever to do with biology. One method that was popular about 10 years ago was using computerized compression.
This is based on the notion of complexity — all objects are complex to one degree or another, but the information they contain can often be reduced (or compressed) to a smaller amount without losing anything essential. That is, the original information can be exactly reproduced from the compressed version. This notion is applied to computer files, for example, in the idea of zipping a file — depending on the file's content it may be possible to zip the file to much a smaller size for storage or transmission.
This idea is straightforward to apply to similarity (Cilibrasi & Vitányi 2005). One simply has to compress the two objects separately as well as compress the combination of the two objects (ie. their union or concatenation). If the two objects are identical then the compression of their union will be the same size as the compression of each object individually (= a distance of 0), because all of the information in one of the objects is redundant. If they have nothing in common then the compression of their union will be the sum of the sizes of the compression of each object individually (= a distance of 1).
This idea has been applied in a number of areas where similarity / distance is used to group objects together (ie. clustering or classification), including computer viruses (Wehner 2008), music (Cilibrasi et al. 2004), languages (Benedetto et al. 2002a), and genomics (Chen et al. 2000, Li et al. 2001, Otu et al. 2003).
This approach has not always been received with equanimity. For example, the paper by Benedetto et al. (2002a) has received considerable commentary (Khmelev & Teahan 2003a; Benedetto et al. 2003; Khmelev & Teahan 2003b), (Goodman 2002; Benedetto et al. 2002b), (Wang 2009).
In biology, this idea has been mostly ignored. Chen et al. (2000) and Li et al. (2001) based their idea of information on Kolmogorov complexity, which is "the shortest program on a universal computer that outputs one of the objects when the input is the other object". This complexity cannot be measured directly, and so it is approximated by using a file compression program (eg. gzip). Clearly, the approximation can only be as good as the compression program and its algorithms.
Otu et al. (2003) based their idea of information on Lempel & Ziv complexity, which is "related to the number of steps required by a production process that builds either of the two objects". They claim an exact solution to the measurement of this complexity for gene sequences, and propose several slightly different distances based on this. Furthermore, the claim to:
show that the proposed distance measures, which are based on the relative complexity between sequences imply the evolutionary distance between organisms ... As the proposed approach does not depend on multiple alignments we test the validity of the approach in two ways: we use simulated data to show that the proposed distance measures can reasonably be represented by a tree. We also show the superiority of the proposed method on existing techniques using this simulated data. Secondly, we look at how well the results generated by the proposed method agree with existing phylogenies.The use of generic compression similarity simply demonstrates that phylogeny does produce similarity to some extent. However, phylogeny is based on what we might call a form of "special similarity", which is similarity solely due to the possession of shared derived character states. Other components of similarity, such as the similarity produced by parallelisms, convergences and reversals, do not contribute to an estimate of a phylogeny. Indeed, they will often be positively misleading. A general concept of similarity is inadequate for reconstructing a phylogeny except under the simplest circumstances. This was empirically demonstrated, for example, in liguistics by the results of the Computer-Assisted Stemmatology Challenge, in which the compression method CompLearn came dead last in the primary ranking of the different phylogenetic methods (but did okay in the secondary ranking).
In short, only some aspects of complexity are relevant for phylogenetics, and only some of the information contained in a genome is relevant for measuring phylogenetic similarity. The focus on information as a whole ignores the "bio" in bioinformatics.
References
Benedetto D, Caglioti E, Loreto V (2002a) Language trees and zipping. Physical Review Letters 88: 048702. [arXiv:cond-mat/0108530, 2001]
Benedetto D, Caglioti E, Loreto V (2002b) On J. Goodman’s comment to "Language trees and zipping". arXiv:cond-mat/0203275
Benedetto D, Caglioti E, Loreto V (2003) A reply to the comment by Khmelev and Teahan. Physical Review Letters 90: 089804.
Chen X, Kwong S, Li M (2000) A compression algorithm for DNA sequences and its applications in genome comparison. Proceedings of the Fourth Annual International Conference on Computational Molecular Biology (RECOMB), pp. 107-117. ACM Press, Tokyo.
Cilibrasi R, Vitányi PMB (2005) Clustering by compression. IEEE Transactions on Information Theory 51: 1523-1545.
Cilibrasi R, Vitányi P, de Wolf R (2004) Algorithmic clustering of music based on string compression. Computer Music Journal 28(4): 49-67.
Goodman J (2002) Extended comment on "Language trees and zipping". arXiv:cond-mat/0202383
Khmelev DV, Teahan WJ (2003a) Comment on "Language trees and zipping". Physical Review Letters 90: 089803.
Khmelev DV, Teahan WJ (2003b) Comment on the reply of Benedetto et al. arXiv:cond-mat/0303261
Li M, Badger JH, Chen X, Kwong S, Kearney P, Zhang H (2001) An information-based sequence distance and its application to whole mitochondrial genome phylogeny. Bioinformatics 17: 149-154.
Otu HH, Sayood K (2003) A new sequence distance measure for phylogenetic tree construction. Bioinformatics 19: 2122-2130.
Wang X-L (2009) Comment on "Language trees and zipping". arXiv:0903.3669
Wehner S (2008) Analyzing worms and network traffic using compression. Journal of Computer Security 15: 303-320. (arXiv:cs/0504045v1, 2007)
Labels:
Bioinformatics,
Complexity,
distance measure
Monday, May 12, 2014
Automated natural language processing
Natural language processing is all about getting computers to automatically extract information from natural (human) languages, rather than from specially designed computer languages, or even from mathematical datasets.
Each year the Conference on Computational Natural Language Learning (CoNLL) features a practical task, in which participants train and test their own language-parsing systems on exactly the same natural-language datasets. For the tenth CoNLL (CoNLL-X), in 2006, the task was Dependency Parsing. (Previous tasks had included chunking, clause identification, named entity recognition, and semantic role labeling.)
Parsing refers to identifying the words, their associated part of speech (noun, verb, etc) and their syntactic relations (subject, predicate, etc) based on the formal rules of grammar. In computational linguistics the result is often represented as a tree diagram showing the relationships among the words. From this tree we can try to understand the exact meaning of the text. Wikipedia, of course, has an article with more details, if you are interested.
For the 2006 CoNNL, the 18 parsing algorithms were tested using treebanks for 12 different languages. In linguistics, a treebank is a previously parsed body of text with the syntactic or semantic sentence structure annotated. So, the idea is to use some existing treebanks (produced by hand) to train the parsers, and then test them on some new treebanks, to see if they can produce the correct tree. In particular, the testing in 2006 involved what is called dependency grammar, which gives primacy to the verb as the structural center of a clause.
The paper by Buchholz and Marsi (2006) discusses the treebanks for the 12 languages, describes how they were converted into the same dependency format, and provides an overview of the parsing approaches taken by the 18 participants. The methods are named after the first author of the associated paper.
I analyzed the results using a couple of phylogenetic networks. As usual, I used the manhattan distance to evaluate the multivariate relationships in the data, and displayed this using a NeighborNet.
The first graph shows the relationships among the different parsing methods. Methods near each other in the network have a similar parsing success, while methods further apart are progressively more different from each other.
The methods form a simple gradient of increasing average success, from top-left to bottom-right. This means that the methods do not vary much in their success from language to language — if they are successful at parsing one language then they are successful on the other languages as well, and if not then not.
Perhaps this is not unexpected. However, the two most successful methods, by McDonald and Nivre, have quite different approaches to parsing — they differ on nine of the ten characteristics listed by Buchholz and Marsi (2006). Their very similar success is therefore noteworthy — there is apparently more than one way of skinning this particular cat.
The second graph shows the relationships among the different languages used. Languages near each other in the network have a similar parsing success, while languages further apart are progressively more different from each other.
The languages also form a simple gradient of increasing average success, from top-right to bottom-left. The average success at parsing Japanese was 86% (range 65-92%) and the average success at parsing Turkish was 56% (range 38-66%). This does not necessarily mean that Japanese is generally easier to pars than Arabic, Slovene and Turkish, because the datasets themselves varied considerably in the type of text contained in their treebanks. Nevertheless, Arabic, Slovene and Turkish are all "morphologically rich" languages, and parsing them is expected to be hard. It is interesting to note that Dutch is different from the other Germanic languages (Danish, German and Swedish), and Spanish is different from Portugese.
The practical task for the 2014 conference will be Grammatical Error Correction, which was also the task for 2011–2013. The parsers will be given short English texts written by non-native speakers of English, and they will be evaluated on their ability to detect the grammatical errors and provide corrected texts. English is an ideal language for this task, as it is often suggested that for every native speaker of English there are 4–5 non-native speakers, and therefore automated correction of text would be of enormous practical value. (Mandarin Chinese has more speakers in total, but most of these are native speakers.)
Reference
Buchholz, Sabine and Marsi, Erwin (2006) CoNLL-X shared task on multilingual dependency parsing. In: Proceedings of the 10th Conference on Computational Natural Language Learning, pp. 149-164. Association for Computational Linguistics, Stroudsburg, PA.
Labels:
EDA,
Linguistics
Wednesday, May 7, 2014
Lager beer and phylogenetic networks
Beer and wine making have been known since ancient times, but both were usually a hit-or-miss affair. However, beer brewing gradually changed during the Middle Ages in Europe to produce a consistent ale-type beer. The process is carried out by the yeast Saccharomyces cerevisiae, which is also involved in producing wine and leavened bread.
Lager beer, on the other hand, was first brewed in the 15th century in Bavaria (now part of Germany), and it has since become the most popular type of beer worldwide. Lager is bottom fermented (versus top-fermented for ale), and the fermentation is carried out at a lower temperature (< 10 °C versus 15-25 °C). It produces a "smooth, crisp, fruity, and clean" beer as opposed to the "robust, hearty and fruity" ale beer.
Lager beers are made using a yeast known as Saccharomyces pastorianus (also known as Saccharomyces carlsbergensis). This is a domesticated species that depends on humans for its propagation, and it is an allotetraploid hybrid. For a long time, we have known that half of the genome came from the ale yeast Saccharomyces cerevisiae, but the other half came from an unidentified species. In 2011, Libkind et al. (Microbe domestication and the identification of the wild genetic stock of lager-brewing yeast. Proceedings of the National Academy of Sciences of the USA 108: 14539-14544) identified the missing parent yeast as the previously unknown species Saccharomyces eubayanus, which grows on Nothofagus (Southern Beech) trees in Patagonia (in Argentina).
Lager was developed when the Bavarians started to brew and store their beer in caves or cellars, which kept it at a constant cool temperature. This required a yeast that could tolerate the colder temperatures, and it is apparently this component that Saccharomyces eubayanus contributes to the Saccharomyces pastorianus hybrid. Precisely how it got from Patagonia, at the southern tip of South America, to south-eastern Germany is not clear, but perhaps some were carried on the feet of fruit-flies.
Since we are dealing with an allotetraploid hybrid, and thus reticulate evolution, a phylogenetic network would be the appropriate means to illustrate its origins. This has recently been provided by Peris et al. (2014. Population structure and reticulate evolution of Saccharomyces eubayanus and its lager-brewing hybrids. Molecular Ecology 23: 2031-2045). They report that:
genetically diverse strains of S. eubayanus are readily isolated from Patagonia, demonstrating that the species is well established there. Analyses of multilocus sequence data strongly suggest that there are two diverse and highly differentiated Patagonian populations. The low nucleotide diversity found in the S. eubayanus moiety of hybrid European brewing strains suggests that their alleles were drawn from a small subpopulation that is closely related to one of the Patagonian populations ... These findings show how genetically diverse eukaryotic microbes can produce rare but economically important hybrids with low genetic diversity when they migrate from their natural ecological context.
The authors actually present two networks, the first of which is shown here. It is an unrooted supernetwork derived from the the maximum-likelihood trees of each of eight nuclear genes. It shows the network of samples from three named Saccharomyces species, along with expanded views of the two Patagonian populations. Isolates W34/70 and BCS1503 are from the lager yeast, Saccharomyces pastorianus, which in the network are related to the Patagonia B population. The three Saccharomyces bayanus samples (beer contaminants) are hybrids between Saccharomyces eubayanus and Saccharomyces uvarum.
Labels:
Beer,
SuperNetwork
Monday, May 5, 2014
Word cloud of publications on phylogenetic networks
There are a number of ways available to analyze word frequency and usage in a block of text, and to display the result as a diagram. Word clouds, for example, use font size in the diagram to represent word frequency in the text.
I thought that it would be interesting to look at the recurring words in the abstracts of papers about phylogenetic networks. So, I used the search phrase "phylogenetic network" to obtain the relevant abstracts indexed in PubMed, using the ebot server to produce a perl script that performed the search, and then used the Wordle server to generate a word cloud. The search produced 1,285 publications, from which I deleted the irrelevant PubMed-produced information before producing the diagram. (I also deleted common words in authors' addresses.)
Many of the words are not very revealing about the subject. Nevertheless, I conclude from this that more of the papers involve gene sequences of different species, while population studies less often refer to their display diagrams as "phylogenetic networks", particularly for studies of mitochondrial data (they probably use "haplotype network", instead). Other than that, the only organism in the diagram is "human". "Phylogeny", "tree/s" and "clade" all make an appearance, revealing the strong link to the past.
I thought that it would be interesting to look at the recurring words in the abstracts of papers about phylogenetic networks. So, I used the search phrase "phylogenetic network" to obtain the relevant abstracts indexed in PubMed, using the ebot server to produce a perl script that performed the search, and then used the Wordle server to generate a word cloud. The search produced 1,285 publications, from which I deleted the irrelevant PubMed-produced information before producing the diagram. (I also deleted common words in authors' addresses.)
Many of the words are not very revealing about the subject. Nevertheless, I conclude from this that more of the papers involve gene sequences of different species, while population studies less often refer to their display diagrams as "phylogenetic networks", particularly for studies of mitochondrial data (they probably use "haplotype network", instead). Other than that, the only organism in the diagram is "human". "Phylogeny", "tree/s" and "clade" all make an appearance, revealing the strong link to the past.
Labels:
Wordle
Subscribe to:
Comments (Atom)