These days, there are many unrooted affinity-type networks used to display conflicting phylogenetic signals. There are many different methods available, although the various forms of splits graphs seem to dominate, especially NeighborNet and Consensus Networks (for species-level data), and Reduced Median Networks and Median Joining Networks (for population-level data). However, phylogeneticists are interested in genealogies, not just data displays.
Unfortunately, rooted evolutionary networks are not so well off. There is a great need for such networks in phylogenetics, but there are very few automated methods available for constructing them. These networks are needed whenever a genealogy involves reticulation processes rather than solely divergence. The latter produces a tree-like evolutionary history but the former do not, and these thus require network methods.
Due to the lack of obvious methods, most current research papers still do not illustrate reticulate evolution with a genealogy. A collection of ad hoc methods is usually applied to the data, and the evolutionary processes are then inferred from this. However, the use of a network to illustrate the inferred genealogy is rather rare.
Indeed, for species-level studies most papers simply present a set of incongruent gene trees, although some of them also illustrate either (i) the tree derived from the combined data, or (ii) a consensus tree with or without the conflicting relationships, or (iii) a pair of cophylogeny trees. Occasionally, the hybrid origin of some of the species, for example, is illustrated, but the putative parents are not connected in a phylogeny.
Population-level studies often present unrooted haplotype networks, illustrating processes such as hybridization and introgression between closely related species, or the evolution of domesticated species.
However, these ad hoc methods do not mean that evolutionary networks do not appear in the literature. In this blog post I include a representative sample of rooted networks that are intended to illustrate inferred genealogies. They are grouped according to the evolutionary processes being studied (see Reticulation patterns and processes in phylogenetic networks). I have also briefly indicated how the networks were constructed.
Homoploid Hybridization
Hybridization is commonly studied in the literature, and phylogenetic networks appear not infrequently. This first example was constructed by the unreleased program HyperPars.
Dickerman AW (1998) Generalizing phylogenetic parsimony from the tree to the forest. Systematic Biology 47: 414-426.
This next example was constructed by program SplitsTree. Note that the root of the network is not clearly indicated.
Pirie MD, Humphreys AM, Barker NP, Linder HP (2009) Reticulation, data combination, and inferring evolutionary history: an example from Danthonioideae (Poaceae). Systematic Biology 58: 612-628.
This example was constructed manually from a set of gene trees. Note that it is drawn in a rather unusual style for indicating hybridization.
Sang T, Crawford D, Stuessy T (1997) Chloroplast DNA phylogeny, reticulate evolution, and biogeography of Paeonia (Paeoniaceae). American Journal of Botany 84: 1120-1136.
Polyploid Hybridization
Polyploid hybridization is probably the most likely type of study to have a phylogenetic network. This is at least partly because there is a computer program, Padre, to automate much of the work. This program was used to construct this first network.
Marcussen T, Jakobsen KS, Danihelka J, Ballard HE, Blaxland K, Brysting AK, Oxelman B (2012) Inferring species networks from gene trees in high-polyploid North American and Hawaiian violets (Viola, Violaceae). Systematic Biology 61: 107-126.
This next example was also constructed by program Padre.
Sessa EB, Zimmer EA, Givnish TJ (2012) Unraveling reticulate evolution in North American Dryopteris (Dryopteridaceae). BMC Evolutionary Biology 12: 104.
This example constructed manually from a gene tree.
Marhold K, Lihová J (2006) Polyploidy, hybridization and reticulate evolution: lessons from the Brassicaceae. Plant Systematics and Evolution 259: 143-174.
Introgressive Hybridization
Introgression is a widely studied phenomenon. However, rooted evolutionary networks are rarely presented. This first one was constructed manually from a set of gene trees.
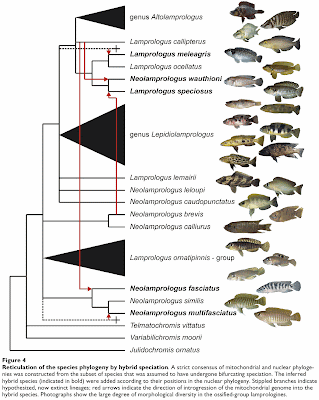
Koblmüller S, Duftner N, Sefc KM, Aibara M, Stipacek M, Blanc M, Egger B, Sturmbauer C (2007) Reticulate phylogeny of gastropod-shell-breeding cichlids from Lake Tanganyika — the result of repeated introgressive hybridization. BMC Evolutionary Biology 7: 7.
The next example was also constructed manually from a set of gene trees.
Morgan DR (2003) nrDNA external transcribed spacer (ETS) sequence data, reticulate evolution, and the systematics of Machaeranthera (Asteraceae). Systematic Botany 28: 179-190.
This example was constructed by program SplitsTree.
Labate JA, Robertson LD (2012) Evidence of cryptic introgression in tomato (Solanum lycopersicum L.) based on wild tomato species alleles. BMC Plant Biology 12: 133.
Horizontal Gene Transfer
HGT is a hot topic these days, both among prokaryotes and among eukaryotes, although most papers do not present a phylogenetic network. The first example was constructed by program Sprit from the species tree and a gene tree.
Walsh AM, Kortschak RD, Gardner MG, Bertozzi T, Adelson DL (2013) Widespread horizontal transfer of retrotransposons. Proceedings of the National Academy of Sciences USA 110: 1012-1016.
This next example was constructed manually from a gene tree.
Delwiche CF, Palmer JD (1996) Rampant horizontal transfer and duplication of rubisco genes in eubacteria and plastids. Molecular Biology and Evolution 13: 873-882.
This example was constructed manually from incongruence among a series of gene trees.
Richards TA, Soanes DM, Foster PG, Leonard G, Thornton CR, Talbot NJ (2009) Phylogenomic analysis demonstrates a pattern of rare and ancient horizontal gene transfer between plants and fungi. The Plant Cell 21: 1897-1911.
Homologous Recombination
Intra-genic recombination is often studied without reference to a network. Nevertheless, several programs exist, and this particular network was constructed by program Kwarg.
Jenkins PA, Song YS, Brem RB (2012) Genealogy-based methods for inference of historical recombination and gene flow and their application in Saccharomyces cerevisiae. PLoS One 7: e46947.
Chromosomal rearrangements are studied rather rarely. This network was constructed manually from a phylogenetic tree. Note that the root of the network is not clearly indicated.
Rumpler Y, Hauwy M, Fausser JL, Roos C, Zaramody A, Andriaholinirina N, Zinner D (2011) Comparing chromosomal and mitochondrial phylogenies of the Indriidae (Primates, Lemuriformes). Chromosome Research 19: 209-224.
Viral Reassortment
Reassortment of segmented viruses produces very complex networks. This one is a partial network, constructed manually from a series of phylogenetic analyses.
Smith GJ, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, Peiris JS, Guan Y, Rambaut A (2009) Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459(7250): 1122-1125.
Genome Fusion
This is a difficult topic to study. As is almost always done, this network was constructed manually from a phylogenetic tree.
Thiergart T, Landan G, Schenk M, Dagan T, Martin WF (2012) An evolutionary network of genes present in the eukaryote common ancestor polls genomes on eukaryotic and mitochondrial origin. Genome Biology and Evolution 4: 466-485.
Apomixis
This topic rarely involves networks. This network was constructed manually from the output of program SplitsTree.
Dyer RJ, Savolainen V, Schneider H (2012) Apomixis and reticulate evolution in the Asplenium monanthes fern complex. Annals of Botany 110: 1515-1529.
Removing Convergence
This is an unusual use of a network, but the author notes that "the use of reticulations clarifies the phylogeny by factoring out apparent convergence, even though there is no reason to think that actual hybridization or introgression has occurred." The network was constructed by an unreleased program.
Alroy J (1995) Continuous track analysis: a new phylogenetic and biogeographic method. Systematic Biology 44: 152-178.


















This is a really important blog posting because it collects many important examples of evolutionary network use together in one place. You should make an article out of this David...then we can all cite it in the introduction to our articles. Will save us a lot of work ;)
ReplyDeleteI am glad that you like it, since it was actually written specifically with you in mind. It follows from some of our email exchanges, which made it clear to me that a summary is needed (by me, if nobody else!). A paper would probably require combining this and the previous post on "Reticulation patterns", I guess.
DeleteCould you direct us to the "Reticulation patterns" post?
ReplyDeleteThanks.
Steve
It is in the text above. It is:
Deletehttp://phylonetworks.blogspot.com/2013/10/reticulation-patterns-and-processes-in.html