The term "DNA barcoding" is a metaphor, and like all metaphors it is helpful only to the extent that it provides insight into the topic at hand. The metaphor concerns commercial barcodes, which were developed to provide a means of storing and retrieving information about manufactured products. Once a product exists we can create a barcode that uniquely identifies that product. At any future time we can invert this chain of logic, by reading the barcode and thus retrieving information about the product.
Metaphor
Does this metaphor apply in the biological world? Well, partly. Whenever biological variation is discontinuous then we could treat the delimited entities as analogous to products, and some part of the DNA must be unique and could be used as a unique identifier. However, much biological variation is more or less continuous, and at best delimits fuzzy (ie. overlapping) clusters rather than discrete entities; so even the theoretical idea that we could know about biodiversity by reading barcodes is not a forgone conclusion.
Just as importantly, however, barcodes apply to one part of the genome, while biodiversity is about whole organisms and their relationships. Barcodes do not apply to either genomes or organisms, they apply to genes. How many barcodes does a genome need before it is uniquely characterized? A product needs only one, but that is because we defined the product first and then applied the barcode to it. But in biology we read the barcode first and then try to work out what it might apply to.
Furthermore, does barcoding a genome also barcode the organisms? Not that we know of. Each organism is a phenotype, which is a genotype interacting with its environment (in the broadest sense). There is much more to biodiversity then merely a collection of genomes. So, even if we do have a DNA barcode, we don't really know what this tells us about biodiversity.
So, a DNA barcode provides information but not necessarily knowledge, whereas a product barcode provides both. Therein lies the major weakness of the metaphor.
DNA barcoding seems to have started as a means of identifying DNA in foodstuffs, and in this application the metaphor seems to have some use, because the weakness does not have much affect. After all, we are mainly trying to identify DNA that is foreign to the alleged ingredients, which merely asks the question: Is there more to this food item than meets the eye? Since the ingredients are all distinct entities, and we know about them beforehand, all we are doing is identifying the entities by examining their barcodes.
However, DNA barcoding is now being used to help create a catalogue of life, which is a completely different thing. In this application, we are trying to delimit entities based on their alleged barcodes — if they have different barcodes then they thus must be different entities. We are counting barcodes but we are not necessarily counting meaningful biological entities. Here, the metaphorical weakness seems like a major handicap, potentially leading to mis-interpretation of what DNA barcoding can and cannot achieve.
DNA barcoding is a viable technology for helping to quantify DNA diversity, which is what it is used for when examining foodstuffs. But the metaphor should not lead us to the conclusion that information about DNA diversity automatically provides much knowledge about biodiversity as a whole. We would end up with a catalogue, but we would not necessarily know what it refers to. This would be a data-base but it would not be a knowledge-base.
Networks
What does this have to do with phylogenetic networks? Well, the criteria for defining entities and identifying them based on DNA barcodes is usually a phylogenetic tree. We create a phylogenetic tree of the known barcodes, and the closest barcode in the tree is then used as the best "identification" of any newly discovered barcode. Remember, product barcodes are unique by definition, and we know what they refer to. But DNA barcodes are not unique unless we decide that they are; and we have no prior idea what they refer to. We make both decisions with reference to clades on a phylogenetic tree.
But a phylogenetic tree imposes a hierarchical structure on the data, irrespective of whether there actually is such a structure underlying the data. A phylogenetic network might reveal a very different pattern. In particular, when the data are forced into a tree then many of the shared characters become parallelisms and reversals, whereas the network can actually display them as shared characters.
To illustrate this, we can look at some of the data from the first published paper about DNA barcodes:
Hebert PD, Cywinska A, Ball SL, deWaard JR (2003) Biological identifications through DNA barcodes. Proceedings of the Royal Society of London B: Biological Sciences 270: 313-321.The authors evaluated the usefulness of cytochrome c oxidase I (COI or Cox1) sequences as a barcode. They analyzed sequences 223 amino-acids long from 100 members of the Bilateria. The original analysis was based on Poisson-corrected p-distances and the Neighbour-joining algorithm — chosen because of "its strong track record in the analysis of large species assemblages [and] the additional advantage of generating results much more quickly than alternatives." The tree was shown as rooted on the Platyhelminthes but without explanation (the other two analyses in the same paper had clearly specified outgroups). The tree itself looks like it might have a mid-point root.
No measure of branch support was provided, but the authors concluded that their analysis:
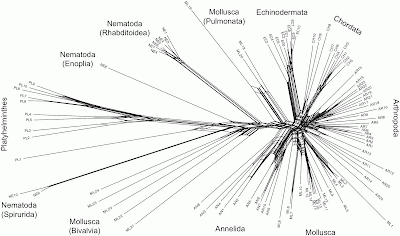
showed good resolution of the major taxonomic groups. Monophyletic assemblages were recovered for three phyla (Annelida, Echinodermata, Platyhelminthes) and the chordate lineages formed a cohesive group. Members of the Nematoda were separated into three groups, but each corresponded to one of the three subclasses that comprise this phylum. Twenty-three out of the 25 arthropods formed a monophyletic group, but the sole representatives of two crustacean classes (Cephalocarida, Maxillopoda) fell outside this group. Twelve out of the 25 molluscan lineages formed a monophyletic assemblage allied to the annelids, but the others were separated into groups that showed marked genetic divergence. One group consisted solely of cephalopods, a second was largely pulmonates and the rest were bivalves.I have tried to reconstruct the data (it is not available online), and re-analyzed it using Neighbor-Net (the closest network equivalent of Neighbour-joining) and uncorrected p-distances.
Some of the recognized taxonomic groups are, indeed, characterized by splits in the network, notably the Echinodermata, the Annelida, the Pulmonata (Mollusca), and the various parts of the Nematoda. However, the other groups are ambiguously defined. In particular, the Chordata, Arthropoda and most of the Mollusca are indistinct based on the gene sequence being analyzed, and there is no split supporting the Bivalvia (Mollusca). There is a split supporting the Platyhelminthes, but it has strong reticulate relationships with parts of the Nematoda — this is unfortunate since this is allegedly the root. Removing sample PL1 from the analysis makes the root a bit less ambiguous, and the network then unites most of the Nematoda as a single group.
This network does not really support the methodology used by the original authors. The authors tested the viability of DNA barcoding by adding a series of "test" sequences, one at a time, to the tree-based analysis, to see whether these sequences clustered with the "correct" group in the tree. However, most of the sequences don't form clear groups in the network, so it is not obvious how one would unambiguously decide which alleged group each test sequence clusters with.
The barcode metaphor looks very poor in this network. I wonder whether DNA barcoding would have taken off if the authors had presented this network rather than their original tree?


No comments:
Post a Comment