I have previously noted that splits graphs are a logical way to present the results of Bayesian analyses (We should present bayesian phylogenetic analyses using networks). Bayesian analyses are concerned with estimating a whole probability distribution, rather than producing a single estimate of the maximum probability. Thus, the result of a Bayesian phylogenetic analysis should not be as a single tree (the so-called MAP tree or maximum a posteriori probability tree), but should instead show the probability distribution of all of the sampled trees. This can easily be done with a consensus network, as illustrated by example in the previous blog post.
An interesting alternative way of visualizing the probability distribution of trees is what has been called a Cloudogram, an idea introduced by Remco R. Bouckaert (2010, DensiTree: making sense of sets of phylogenetic trees. Bioinformatics 26: 1372-1373). This diagram superimposes the set of all trees arising from an analysis. Dark areas in such a diagram will be those parts where many of the trees agree on the topology, while lighter areas will indicate disagreement. This idea can be best illustrated by a few published examples.
The first cloudogram is from Figure 4 of Chaves JA, Smith TB (2011) Evolutionary patterns of diversification in the Andean hummingbird genus Adelomyia. Molecular Phylogenetics and Evolution 60: 207-218.
In this case the MAP tree has been superimposed on the cloudogram.
The next one is from Figure 2 of Pabijan M, Crottini A, Reckwell D, Irisarri I, Hauswaldt JS, Vences M (2012) A multigene species tree for Western Mediterranean painted frogs (Discoglossus). Molecular Phylogenetics and Evolution 64: 690-696.
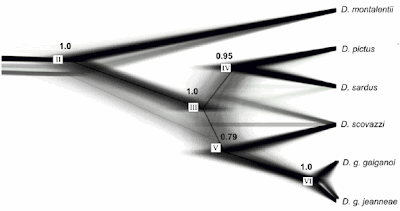
The next one is from Figure 1 of Lerner HR, Meyer M, James HF, Hofreiter M, Fleischer RC (2011) Multilocus resolution of phylogeny and timescale in the extant adaptive radiation of Hawaiian honeycreepers. Current Biology 21: 1838-1844.
In this case the data are more tree-like than the previous two examples.
The final one is from Figure 2 of McCormack JE, Faircloth BC, Crawford NG, Gowaty PA, Brumfield RT, Glenn TC (2012) Ultraconserved elements are novel phylogenomic markers that resolve placental mammal phylogeny when combined with species-tree analysis. Genome Research 22: 746-754.
This analysis involves bootstraps rather than Bayesian samples, showing that the same principle applies.
It would be nice to illustrate this further by direct comparison with a splits graph of the same dataset that I used in the previous blog post. Unfortunately, the computer program available (DensiTree) has the same practical limitation as the SplitsTree program (as mentioned in the previous post) — it does not read the MrBayes ".trprobs" file because it ignores the tree weights. This means that one has to enter the entire treefile (with thousands of trees), and I have not yet done that. Moreover, the program relies very much on having branch lengths for each tree — the output is really quite odd without them, with the taxa appearing in a series of steps rather than connected by straight branches. My previous analysis did not use branch lengths, as they are not needed for the consensus network, in which edge lengths represent support rather than character evolution.




Those are great -- would love to see that become the standard.
ReplyDeleteCloudograms could also be a good way to represent discordance in gene trees across loci (e.g. our recent paper that has cloudograms of MCC trees from each of ~160 UCEs: http://sysbio.oxfordjournals.org/content/63/1/83.short). But presenting discordance across loci and probability distributions within loci simultaneously could get messy...
ReplyDeleteIndeed, between-loci variation is a good example of the uses of cloudograms. Also, I agree that trying to represent two different sources of variation in a single cloudogram is likely to be less than helpful !
ReplyDelete