Native speakers of any language will judge the "difficulty" of another language by how much it differs from their own. For example, the Foreign Service Institute (FSI) of the U.S. Department of State lists
five categories of increasing time taken for native English speakers to acquire "General Professional Proficiency" in other languages. This refers to an average, of course, and anyone may personally find one language or another more easy or difficult than others.
FSI Category I (the least time needed) includes most of the Germanic and Romance languages, since English was originally a Germanic language that received a huge Romance input after the Normans turned up in Britain in 1066. The exception is German itself, which is alone in Category II (needing longer), because of its more complex grammar. Category V (the longest time needed for proficiency) consists of Arabic, Cantonese, Japanese, Korean and Mandarin, with Japanese being considered the most difficult.
Most languages are in Category IV, including the rest of the Indo-European languages. The recognizably tougher ones in that group are the Uralic languages (Estonian, Finnish and Hungarian), because of their countless noun cases. Interestingly, Category III (easier than IV) consists of Indonesian, Malaysian and Swahili, which have no known historical connection to English — they just happen to have fewer linguistic differences than do the other languages.
And that is the point of this post — linguistic similarities don't necessarily reflect the evolutionary history of the languages. There
are trees allegedly showing the genealogy of languages, because there is vertical transfer of information in the history of languages (generation to generation), but horizontal transfer has also been a powerful evolutionary force, as cultures come in contact with each other. The history of English, as noted above, shows both vertical (Germanic) and horizontal (Romance) influences. Language history is a reticulating network, not an evolutionary tree.
Just as importantly, though, languages can have coincidental similarities. There are, after all, not that many different ways of constructing a language, and there are reported to be ~6,900 distinct languages on this planet. So, chance similarities must abound — what in biology we would call parallelisms and convergences. This makes constructing the evolutionary history of languages difficult.
The complexity created by coincidences has lead some people to wonder about how "unusual" any one language might be. This can be defined as how many of its characteristics occur commonly in other languages, and how many of them occur more rarely. The most unusual languages will be those that have lots of the rare features; and we might call them linguistic outliers. The Idibon blog has already had a look at this topic (
The weirdest languages), and here I reconsider their data in the light of a phylogenetic network.
The data
The original data come from the
World Atlas of Language Structures, which describes itself as "a large database of structural (phonological, grammatical, lexical) properties of languages gathered by a team of 55 authors". There are apparently 2,676 different languages in the database, coded for 192 linguistic features. Sadly, the database is very sparse, so that most languages have not yet been coded for most of the features (there are 5–1,519 languages coded for each feature).
So, the Idibon people selected a subset of the data: 1,693 languages and 21 features. These features were chosen to be an uncorrelated subset of those 165 features that have at least 100 languages coded; and the selected languages each have at least 10 features coded.
The features are certainly an eclectic collection, which you can read about on the
WALS site:
83A:
87A:
143A:
143G:
69A:
116A:
57A:
101A:
6A:
71A:
129A:
130A:
44A:
14A:
9A:
72A:
111A:
64A:
124A:
117A:
19A:
|
Order of Object and Verb
Order of Adjective and Noun
Order of Negative Morpheme and Verb
Minor Morphological Means of Signaling Negation
Position of Tense-Aspect Affixes
Polar Questions
Position of Pronominal Possessive Affixes
Expression of Pronominal Subjects
Uvular Consonants
The Prohibitive
Hand and Arm
Finger and Hand
Gender Distinctions in Independent Personal Pronouns
Fixed Stress Locations
The Velar Nasal
Imperative-Hortative Systems
Nonperiphrastic Causative Constructions
Nominal and Verbal Conjunction
'Want' Complement Subjects
Predicative Possession
Presence of Uncommon Consonants |
From the subset of languages, I chose all of those languages with at least 12 of these features coded, plus Icelandic (10 features), and Cornish and Gaelic(Scots) (11 features).
I then tried to fill in some of the missing data, to get as many languages as easily possible up to having 14 features coded (ie. two-thirds of the features). For the phonology features (6A, 9A, 19A), the relevant information can be looked up on the web, particularly in Wikipedia and the
Native American Language Net. For the word features (129A, 130A), I used the
LEXILOGOS Online Translation.
In the process, I found that Idibon has at least one feature mis-coded compared to the WALS web site: for feature 14A, some of the languages that should be coded "Second " have been coded as "Antepenultimate", and all of the others that should be coded "Second" have missing data.
I also found a few contradictions between the WALS coding and the information elsewhere on the web. In some of these cases I re-coded the WALS data.
My final spreadsheet is available
online. There are 280 languages coded for at least 14 of the 21 features, compared to 239 such languages in the Idibon analysis. There are 19% of the data still missing, varying from 0–53% across the 21 features.
The network
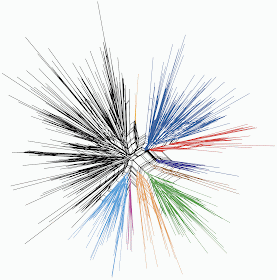
My network is intended as an exploratory data analysis, rather than some attempt at an evolutionary diagram. Thus, the network simply displays the apparent similarity among the languages. That is, languages that are closely connected in the network are similar to each other based on their linguistic features, and those that are further apart are progressively more different from each other.
First, I recoded the multivariate linguistic data as 59 binary characters. Then the similarity among the 280 languages was calculated for each pair of languages using the Gower similarity index, which can accommodate missing data (by ignoring features that are missing for each pairwise comparison). A Neighbor-net analysis was then used to display the between-language similarities as a phylogenetic network.
The network is not very tree-like, is it? A few tentative groups can be recognized, as indicated by my colouring, but that is all. These groups do not correspond to any known language groups, meaning that the language features chosen do not reveal a traditional tree-like genealogy. Whether this reflects horizontal transfer of linguistic features, coincidence, or simply inadequate data, is not necessarily clear.
However, it seems most likely that much of the complexity represents coincidence. In the study of language evolution, parallelism and convergence are not nuisances, which is the way they are treated when constructing phylogenies of organisms. Coincidental similarities are a fundamental part of language history, but they are not necessarily the product of processes like natural selection, as they often are in biology.
If we look at some of the details, the nature of the complexity becomes clearer, as shown in the next figure. Here, I have colour-coded the Indo-European family of languages by their so-called "genus", plus the other languages that occur in Europe (the Uralic group, and Basque):
Albanian - pale brown
Armenian - dark brown
Baltic - orange
|
Celtic - pale blue
Germanic - black
Greek - pale green
|
Indic - pink
Iranian - blue
Romance - purple
|
Slavic - green
Uralic - red
Basque - grey
|
Note that the seven Germanic languages are clustered in a single location, as are the two Baltic languages. The others appear in either two (Celtic, Romance, Iranian) or four (Indic, Slavic, Uralic) locations. This implies considerable linguistic variation within most of what are considered to be closely related languages (that is why they are called language genera). A larger collection of features might change the pattern, of course, but I still reckon that there is a large component of non-vertical transmission here. This is either coincidence or horizontal transmission. For the Indo-European languages, the latter is perhaps quite likely; but it is equally likely that it is simply coincidence, even at this relatively fine scale.
The weirdest languages
The Idibon blog tried to reduce the multivariate data down to a single number for each language (scaled 0–1), representing its "weirdness" in terms of how many uncommon features it has. So, I have performed the same calculation for my expanded dataset.
The complete list is in the
spreadsheet, but here are the top and bottom most-unusual languages:
Top 20
Mixtec (Chalcatongo)
Seri
Nenets
Diegueño (Mesa Grande)
Oromo (Harar)
Choctaw
Kutenai
Iraqw
Danish
Kongo
Norwegian
Dutch
Swedish
German
Armenian (Eastern)
Abkhaz
Mumuye
Ju|'hoan
Khoekhoe
Ladakhi
|
0.9725
0.9354
0.9346
0.9196
0.9187
0.9138
0.9079
0.9005
0.8843
0.8830
0.8751
0.8705
0.8585
0.8581
0.8473
0.8445
0.8410
0.8346
0.8300
0.8247
|
Bottom 20
Kanuri
Kunama
Kiowa
Marathi
Khanty
Turkish
Bulgarian
Wichita
Manam
Kewa
Sentani
Bororo
Usan
Cantonese
Hungarian
Chamorro
Ainu
Cherokee
Purépecha
Hindi
|
0.2410
0.2401
0.2361
0.2752
0.2149
0.2145
0.2112
0.2054
0.2085
0.1984
0.1952
0.1534
0.1508
0.1435
0.1316
0.1285
0.1277
0.1232
0.0997
0.0872
|
My results differ from those of the Idibon blog for two reasons: more languages, and more data for some of the languages. Some of my added languages make it to the top of the weirdness list, including Seri, Danish and Swedish; and some of the other languages considerably change their score — for example, Hebrew, Welsh, Portuguese and Chechen are now near top of the list, and Quechua, Basque, Saami and Cornish are no longer near bottom. All of the big changes are increases in weirdness, suggesting that the missing data are important for this calculation.
Nevertheless, it is worth noting that five of the seven Germanic languages are in the top 15 (plus English is at 40 and Icelandic 47). Unusually, most of the Germanic languages still use cases (modifications to words that show how they relate to other words in a sentence). This means that you have to memorize a lot of different versions of each noun, just as you do in Latin. Moreover, these languages change the word order when asking a question as opposed to making a statement, whereas most languages add a participle instead. (In the most unusual language,
Mixtec, a native language from Mexico, there is apparently no difference between a question and statement!)
English has a lower score than other Germanic languages presumably because of the French influence mentioned above (French is ranked 42). For example, in English there are now very few cases (only for some pronouns), as in the other Germanic languages, but instead it uses a fairly strict word order to express grammatical relationships. (You will note that two of the English-speaking authors of this blog now live in countries with other Germanic languages, and so we know just how big a pain it is to learn illogical case endings.)
English does have one really odd feature, though, which is the use of the sound "th" (which is part of feature 19A). There are two forms of this sound, voiced (as in "the") and unvoiced (as in "thing"). These sounds do not exist in most languages, and they are rare even among the other Indo-European languages. That is why you often hear non-native speakers say "dis" and "zis" instead of "this" — "th" is a sound that they have no experience making.
Actually, the Indo-European languages are very diverse in their weirdness. Many of them are at the top of the list, but there are also some at the bottom, including Hindi which is dead last. Notably, three of the Romance languages are at the top (Spanish, Portuguese, French) and two are at the bottom (Romanian, Italian). This seems unlikely, given the overall similarity of Spanish and Italian, for example; and so it probably reflects the specific choice of linguistic features.
The data are also potentially sensitive to some of the feature coding. One notable example is for feature 19A in Arabic.
WALS codes Arabic as having pharyngeals but not "th", while
Wikipedia says that the pharyngeals are doubtful, but that Arabic has "th". So, the possble codings of Arabic, and their resulting weirdness, are:
Feature
"Th" sounds only
Pharyngeals only
Pharyngeals and "th"
|
Score
0.0893
0.0469
0.0045
|
Weirdness
0.6788
0.7416
0.9245 |
So, this feature alone can potentially change Arabic from "normal" to "very weird", depending on how it is coded.
Conclusion
Languages do not have a tree-like evolutionary history. Even the relatively small dataset presented here seems to show the influence of horizontal evolution. But, more importantly, we should not underestimate the coincidental occurrence of language features (parallelism and convergence). These have usually been treated as a nuisance in phylogenetic studies of organisms, but they are likely to be important for the study of languages. I have discussed this further in a previous post (
False analogies between anthropology and biology).

_1894.jpg/179px-Lautrec_confetti_(poster)_1894.jpg)