In my last post, I outlined two ideas about how stacking neighbour-nets can assist in tracing evolutionary change over time, using a theoretical example. In this post, I will show how this could work using a (tricky) real-world example: a morphological matrix including a high proportion of fossil taxa and a good deal of (strongly) homoplasious characters (Bomfleur, Grimm & McLoughlin 2017).
Stacking can be valuable when both fossil and extant taxa are included in the study. The idea of stacking is to construct networks for each time slice, rather than creating one giant network that tries to encompass everything. Adjacent time-slice networks can then be directly compared, which should reveal the evolutionary changes that occurred between those two times. The final phylogeny can then be constructed from this information, including all of the extant taxa and fossils together.
I regard our work as quite innovative for a palaeobotanical/-phylogenetic systematic study, as it generated a taxon-dense dataset down to species (sometimes individual specimens) as ‘operational taxonomic units’ (OTUs). Our goal was to provide a unifying classification for extant and fossil Osmundales (royal ferns) rhizomes. The primary purpose is hence not to infer a phylogenetic tree but to assist in describing and placing new-found rhizome fossils in the phylogeny. The placement workflow (see this tutorial) combines a polytomous key (using conserved, lineage-diagnostic traits) with neighbour-nets that use different taxon sets. We discussed odd placements in the splits graphs, and matrix signal quality (robustness) from differential branch support, as estimated by non-parametric bootstrapping (least-squares, maximum likelihood, maximum parsimony).
Sources of incompatible data patterns in real-world data
The main problem with real-world data when it comes to inferring phylogenetic relationships, i.e. estimating the true phylogeny, are incompatible data patterns. For molecular matrices, the two main sources of signals that will be incompatible with the true phylogeny are back-mutations and model-bias. For instance, there is usually a higher probability for transitions than for transversions; and for coding gene regions, the 3rd codon position can become over-saturated and thus stochastically distributed, providing little phylogenetic signal. By adapting the model in a probabilistic environment, we can (try to) counter such biases during inference
In the case of morphological (or other non-molecular) traits, incompatible signals arise from:
- homoplasious characters – traits that evolve convergently or in parallel, which are frequently included in such matrices;
- epigenetic effects – morphological traits not, or not fully, controlled by the genetic composition of the organism; and
- pseudo-homologies – traits that are seemingly the same but are the endpoint of different evolutionary pathways.
Fossils add further sources of signals incompatible with the true phylogeny, such as: preservation artefacts and misinterpretations (false homologies); uncertainty linked to heterochrony; and, last but not least, ‘temporal’ convergences, i.e. the parallel or convergent evolution of the same (or similar) trait in an ancient sister or unrelated lineage of a modern (or much younger) lineage.
For all of these aspects, the royal fern rhizomes provide a nice example (i.e. a bad-case scenario). Only a few of the 45 scored traits that can be observed in fossil material are conserved within the modern lineages and their extant representatives, and hence are of high diagnostic value for assigning fossils to one of these lineages. Many other rhizome features are variable within extant members of the now six genera (some even within a species), and increasingly so looking back into the past.
The royal ferns became arborescent several times, as reflected by convergent adaptations in rhizome anatomy — highly complex stele architectures are found from the Permian onwards in (morpho)species that differ in all relatively stable, lineage-diagnostic traits. The most complex modern-day rhizomes have anatomies that appear to be less derived than those of some of their ancient counterparts. Nonetheless, the rhizomes, scored for 129/130 OTUs (fossil species, partly referring to individual specimens) in our matrix (click here for an annotated version for use with Mesquite), reflect a substantial past diversity and cover more than 250 million years of evolution.
Basic data situation
The all-inclusive neighbour-net (Fig. 1; see here for a fully annotated version) captures aspects of similarity patterns related to phylogenetic relationships, but does not clearly resolve the known (modern) or putative (extinct) genera within the core group Osmundoideae, for example. Overall branch-support is generally low for any alternative (details can be found here), independent of the optimality criterion used. [For our systematic treatment, we used data subsets to generate a series of networks including only members of the same (putative) lineage, which were increasingly proficient to sort the OTUs.]
The main problems are: (i) the differentiation between less-derived rhizome anatomies of the Osmundoideae found in the likely paraphyletic extinct genus Millerocaulis (pink in Fig. 1) and the modern genus Claytosmunda (magenta, paraphyletic with one survivor); and (ii) the distinctness and superficial similarity of two arborescent lineages, the genus Osmundacaulis (red) and the extinct (Permian to Jurassic) family Guaireaceae (greenish). They differ in all stable, lineage-diagnostic characters but share highly dissected steles. Phylogenetic trees "resolve" this conflict by creating an artificial clade (e.g. the parsimony cladogram by Wang et al. 2014). The neighbour-net (Fig. 1) places Osmundacaulis between the Guaireaceae and the Osmundoideae, the subfamily of Osmundaceae including the surviving modern genera.
 |
| Fig. 1. Neighbour-net based on a morphological distance matrix of 122 OTUs representing Permian to extant Osmundales and their putative relatives, the Grammatopteridales (black). |
Stacking procedure one: identifying closest relatives in subsequent time-slices
Signal ambiguity (from homoplastic characters and the related resolution issue) affects also the time-wise networks to some degree. Figures 2–4 show the network-per-time-slice stacks. Each neighbour-net includes only the OTUs from one stratigraphic period (Permian, Triassic, Jurassic, Cretaceous, Paleogene + Neogene) and the modern-day survivors. For simplicity, links are only established for the closest potential relative in the subsequent or preceding time-slice; and only shown when the mean morphological distance (MD) does not exceed 0.25. The colouring of the dots reflects the systematic affinity of the taxon as established by Bomfleur et al. and shown in Fig. 1.
A major taxonomic turnover characterises the transition from the (late) Permian to the Triassic (Fig. 2). The most primitive (rhizome-wise) Osmundales, the Thamnopterioideae (brown) become extinct, and are completely replaced by the Osmundoideae, their modern counterparts. The only representative of the Permian diversity remaining in the Triassic appears to be Millerocaulis (?Palaeosmunda) stipabonnetiorum, and this may provide a good taxon for rooting the Triassic phylogeny. However, it also one of the worst-preserved and most poorly described taxa — to some degree, its similarity with both lineages of Permian Osmundaceae (Thamnopterioideae and Palaeosmunda) may hint that the distances are under-estimated, since traits could not be scored that otherwise lead to increased distances.
The Jurassic graph (in Fig. 2) highlights a decrease in overall diversity, despite the much higher numbers of OTUs. The links can help to establish relationships between congeners of both time scales; but for Osmundastrum (today represented by a single, genetically and morphologically derived species) a more pronounced evolutionary shift is indicated: the Triassic putative member is linked to Jurassic Millerocaulis species (a paraphyletic Osmundoideae genus defined by the absence of a trait found in all extant genera), which are relatively close to the first unambiguous Osmundastrum. We also find that the three Jurassic newcomers have little relation to the Triassic basis (Fig. 2).
The linking of the Jurassic and Cretaceous time-slices highlights (Fig. 3) a general weakness of the approach using this matrix: poorer preserved, incompletely described fossils included in the matrix (Cretaceous Millerocaulis) attract most links from the Jurassic Osmundoideae — their distances are under-estimated.
The two Osmundastrum, which are probably part of the same evolutionary lineage, are not linked (see Bomfleur, Grimm & McLoughlin 2015 for the reasons). Two modern lineages with more or strongly derived rhizomes appear in the Cretaceous, the Todinae and Plenasium.
In the case of the Todinae the Jurassic links are partly ambiguous, with one Cretaceous OTU linked to Jurassic Claytosmunda (part of the Todinae’s sister clade according to molecular data), but the other with some relatively distinct Millerocaulis. The problem here is that the Todinae may have diverged earlier (Bomfleur, Grimm & McLoughlin 2015; Grimm et al. 2015), but their rhizome fossils have so far not been found (or lack the diagnostic characters of the lineage). Gaps in the fossil record can hinder establishing meaningful links. The links are, however, to a group of Millerocaulis that are closer to coeval Claytosmunda – which show a rhizome anatomy that may be closest to that of the common ancestor of all modern-day king ferns – than to their congeners. In the case of Plenasium, the genus with the most-derived rhizomes of all modern Osmundaceae, the closest older relative is part of the same subgroup of Millerocaulis. These potentially false links may reflect that some Millerocaulis show derived character suites, which are typically found also in one or another modern Osmundaceae genus (similarity due to convergence).
The closer we get to the modern-day situation, the more interpretable the links become (Fig. 4). Lineages with distinct and derived rhizome anatomies such as Osmundastrum and Plenasium are linked across time-slices. Cross-generic links from Cretaceous Millerocaulis to Paleogene-Neogene Osmunda to modern-day Claytosmunda relate directly to higher numbers of shared, possibly primitive characters in the connected taxa; these links can again be informative for rooting the graphs. Substantially weaker links (mean morphological distances > 0.1 between time-slices) are found for distantly related pairings (Cretaceous and extant Todinae with Paleogene-Neogene Osmundastrum and Claytosmunda).
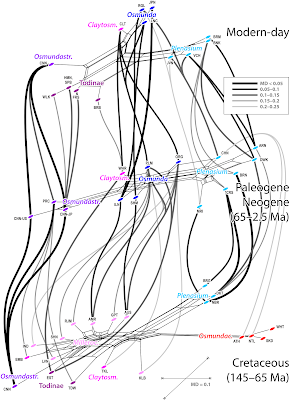 |
| Fig. 4. As above, but for Cretaceous to modern-day. |
Stacking procedure two: graphs including taxa of two subsequent time-slices
Figures 5 and 6 show the two-adjacent-time-slices-per-graph stacks. Interpretation of these figures is more straightforward — one just compares the placement of the connecting taxa (Triassic and Jurassic in Fig. 5; Paleogene and Neogene in Fig. 6). The resolution issue regarding the relationship between Millerocaulis and genera representing the modern lineage (Claytosmunda, Osmundastrum, Plenasium, Leptopteris, Todea) is obvious — the Triassic Millerocaulis are clustered in the Permo-Triassic graph, but are placed apart within the spider-web-like portion in the Triassic-Jurassic graph (Fig. 5). This could mean that several lineages of Millerocaulis diversified in the Jurassic, all of which have their roots in the Triassic. Some of the emerging Millerocaulis groups remain coherent in the Jurassic-Cretaceous graph (and can include Cretaceous species), put their position relative to each other can change. In contrast, for Osmundacaulis the Cretaceous newcomers simply fit into the existing organisation.
The transition from the Cretaceous to the modern-day situation (Fig. 6) fairly reflects what could be inferred by mapping morphological characters onto the molecular tree. The placement of Osmunda species in the graphs reflect evolutionary change towards the modern-day species, whereas stasis can be assumed for Osmundastrum, and a loss of diversity for Claytosmunda. According to the structures of the graphs, the modern-day Plenasium (subgenus Plenasium) replaced the more diverse (and partly more derived) Cretaceous-Paleogene Plenasium (subgenus Aurealcaulis); but the genus is absent from the Neogene, so there are no connections between the ‘65–5 Ma’ and ‘last 25 Ma’ graphs.
 |
| Fig. 6. As above, but covering the time from the Cretaceous to now. Connections refer to Paleogene (lower half) and Neogene (upper half) species. |
Now that it’s done, what can be said?
Establishing similarity links across time-slices can be tedious or even misleading, especially with increasing numbers of taxa and increasing complexity of the signals in the matrix (Figs 2–3). The process is more time-consuming and the result (Figs 2–4) is graphically more challenging than the alternative stacking procedure (Figs 5–6).
With most real-world data, it may be difficult to get a set of links between time slices that reflect the true phylogeny, like it did in my earlier theoretical example. Nonetheless, the procedure can help to identify potential relatives (ancestors, descendants, sister lineages) of groups that are restricted to a single time slice, or highlight the lack of potential or favourable candidates.
However, in general, joining the taxa from two subsequent time-slices in one graph, and connecting these graphs by the shared taxa, seems to be a more feasible and straightforward approach. Once a matrix is compiled, the distance calculation and splits-graph inference is a matter of minutes, and it takes less than half-an-hour to produce a first graphical output using the graphical functions in SplitsTree and software to graphically stack the exported SVG or EPS files (further beautification may take a day). Taxa with odd signals (with ambiguous affinity) will be placed accordingly in the nets and eventually move around in the two containing graphs (Fig. 5) and the amount of evolutionary change across time may be directly visible (Fig. 6).
Additional links for readers interested in details
— Figure illustrating the history of taxonomic systems for Osmundales.
— An archive including all analysis files generated in the course of the original study is hosted at the Dryad Digital Repository.
— Further annotated versions of the figures shown in this post and the used analysis files have been published under a CC-BY licence: Grimm G. (2017) Osmundales diverstity through time: stacking networks. figshare. https://doi.org/10.6084/m9.figshare.5255014.v1.
References
Bomfleur B, Grimm GW, McLoughlin S (2015) Osmunda pulchella sp. nov. from the Jurassic of Sweden—reconciling molecular and fossil evidence in the phylogeny of modern royal ferns (Osmundaceae). BMC Evolutionary Biology 15: 126.
Bomfleur B, Grimm GW, McLoughlin S (2017) The fossil Osmundales (Royal Ferns)—a phylogenetic network analysis, revised taxonomy, and evolutionary classification of anatomically preserved trunks and rhizomes. PeerJ 5: e3433.
Grimm GW, Kapli P, Bomfleur B, McLoughlin S, Renner SS (2015) Using more than the oldest fossils: Dating Osmundaceae with the fossilized birth-death process. Systematic Biology 64: 396-405.
Huson DH, Bryant D (2006) Application of phylogenetic networks in evolutionary studies. Molecular Biology & Evolution 23: 254-267.
Maddison WP, Maddison DR (2001 onwards) Mesquite: a modular system for evolutionary analysis.
Wang S-J, Hilton J, He X-Y, Seyfullah LJ, Shao L (2014) The anatomically preserved Zhongmingella gen. nov. from the Upper Permian of China: evaluating the early evolution and phylogeny of the Osmundales. Journal of Systematic Palaeontology 1: 1-22.




No comments:
Post a Comment