Recently, a number of computer programs have been released that are intended to produce phylogenetic networks representing introgression (or admixture) (see Admixture graphs – evolutionary networks for population biology).
A recent example of the use of these programs is presented by:
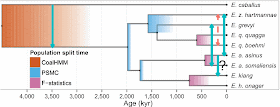
Jónsson H, Schubert M, Seguin-Orlando A, Ginolhac A, Petersen L, Fumagalli M, Albrechtsen A, Petersen B, Korneliussen TS, Vilstrup JT, Lear T, Myka JL, Lundquist J, Miller DC, Alfarhan AH, Alquraishi SA, Al-Rasheid KA, Stagegaard J, Strauss G, Bertelsen MF, Sicheritz-Ponten T, Antczak DF, Bailey E, Nielsen R, Willerslev E, Orlando L (2014) Speciation with gene flow in equids despite extensive chromosomal plasticity. Proceedings of the National Academy of Sciences of the USA 111: 18655-18660.This study presents a phylogenetic analysis of the extant genomes of the genus Equus, the horses, asses and zebras. This analysis leads the authors to the conclusion that there is "evidence for gene flow involving three contemporary equine species despite chromosomal numbers varying from 16 pairs to 31 pairs." The gene flow is indicated by the light-blue reticulations in the first diagram.
One important issue with these types of analyses is the logic on which the procedure is based. Programs like TreeMIx (used in this analysis) were developed to allow modelling of gene flow across the branches of trees at a microevolutionary (population) scale. Specifically, the graph generated by TreeMix models singular (pulse) introgression events in phylogenetic history.
The issue is that a tree is produced first, and then reticulations are added to it. The tree represents descent and the reticulations represent gene flow. But how do we produce a tree from a dataset that contains evidence of both descent and gene flow? The authors' initial tree is shown below.
The procedural logic works as follows:
(i) we assume that the traditionally recognized species exist
(ii) we assume that we have a representative sample of them, with one genome each
(iii) we construct a tree based on the assumption that there is no gene flow among the species
(iv) we then assess the species for gene flow, and discover it.
Isn't this rather circular? Surely (iv) invalidates the assumptions inherent in (i)-(iii)? How can we then assess the reliability of the sampling in (ii) and the analyses in (iii)? Why have we made assumption (i)? At best the species are fuzzy groups to one extent or another, and we do not know where we have sampled within the probabilistic space assigned to the groups.
This seems like a very poor way to go about studying the interaction between descent and gene flow. First we assume descent only, and then we assess gene flow. When we find gene flow we continue to accept the results of the initial analyses based on descent alone.
I would hate to have to justify this philosophy to someone outside phylogenetics, because I have a horrible feeling that they would either smile tolerantly or laugh outright.
This between-species situation is even more extreme for those within-species patterns where groups are recognized. Human races and domesticated breeds are two concepts that have received constant criticism. Neither races nor breeds form clear-cut groups, as there are no sharp boundaries between them, due to gene flow. Their "central locations" in genotype space are usually very different, however. Therefore it is quite possible to perform a tree-based analysis of samples from the central locations, and this would tell us a lot about descent. But it would tell us almost nothing about gene flow; and we would have a very distorted view of the phylogenetic history.


No comments:
Post a Comment