In the book Introduction to Phylogenetic Networks (Morrison 2011), it was convenient to organize the various network types into two groups:
- those that are intended to provide a summary of various possible phylogenetic histories
- those that simply summarize the multivariate data into a convenient visualization.
The consensus network of Holland et al. (2004. Molecular Biology and Evolution 21: 1459-1461) is among the most popular of the networks in the second group. This is formally a Cluster Union Network (CUN), in which the clusters represented by a set of input trees are combined into a single diagram. The clusters are defined by the edges in the original (unrooted) trees - each edge splits the tree into two parts. The trees are thus reduced to the set of splits that appear in at least one of the trees. Each split will then appear in the CUN. If there is no disagreement among the trees, then a split will be represented by a single edge in the CUN; but if there is conflict among the trees then a split will be represented by a set of parallel edges.
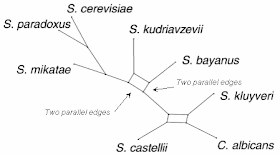 |
| A cluster consensus network, with two reticulation areas, each defined by two sets of parallel edges. |
The end result is that the edges of the CUN no longer represent phylogenetic pathways, even if they did do so in the input trees. Some of the edges of the CUN are there solely as part of a set of parallels. To put it another way, some of the edges do not appear in any of the original trees, but are the result of combining the clusters. So, a CUN will vary from tree-like, if there is little conflict among the input trees (ie. compatible splits) , to a complex spider-web, if there is a lot of conflict (many incompatible splits).
It is this property of representing splits by a set of edges that prevents the network being a representation of phylogenetic history – formally, the edges define clusters not clades.
Miyagi and Wheeler (2019. Cladistics 35: 688-694) have addressed this issue by defining what they call an Edge Union Network. In essence, it is a subset of the CUN - formally, the EUN is contained within the CUN. It can be thought of as a CUN that contains only those edges that appear in at least one of the input trees. M&W see the edges as "redundant" is they appear in the CUN but not the input trees.
M&W's objective for the EUN is thus "to display the total history of all the input trees, rather than the simplest graph which contains all clusters present in the data" (which the CUN does). M&Y see "phylogenetic networks as hypotheses for evolutionary history", so that the EUN can be rooted, just like the input trees. The criterion for the EUN is parsimony, so that "it is important to minimize the number of distinct paths between nodes".
It is important to note in the following discussion that M&W are interested in rooted networks, and so their version of a CUN is not quite the same as the original unrooted Consensus Network.
Discussion
M&W provide a graphical example, the CUN and EUN of two incongruent rooted trees. Here's a colored version: all nodes (internal and terminal) are re-labelled to express the last common ancestor (LCA) that they represent, and internal (conflicting) tree edges are colored, so we can trace them in the networks.
At the bottom left is the strict consensus network, the full CUN, of both trees. Most internal nodes (alternative LCAs) in the trees (ABC, ABCD, AD, DE) are not represented by a single node in the full CUN but by a set of parallel edge bundles (dotted lines). Nonetheless, each edge set represents a branch (clade) in one or both trees – a full CUN depicts all topological alternatives in the two trees. We can extract sets of congruent splits, and reconstruct the two trees in the process.
But since the nodes in the full CUN are not (alternative) LCAs but just connections of (parallel) edges, we cannot interpret this (bottom left) graph as a phylogenetic network. However, the CUN depicted in M&W does do this: we start in the root and walk from node to node along the branches (arrows) until we end up with an explicit phylogenetic network (bottom right). This includes an edge that is not found in any of the trees, a 'false' edge (ABCD-ABC: violet, fat line), while also missing an edge found in one tree (ABCD-BC).
 |
| EUN in comparison to a full CUN for M&W's example. The 'false' ABCD-ABC edge is replaced by a ABCD-BC edge resulting in a phylogenetic network that has only edges seen in the two phylogenetic trees. |
The false ABCD-ABC edge is replaced by an ABCD-BC edge, and ABC is reconnected directly to the root.
An implicit assumption and reason to reduce CUNs into EUNs is that the topological ambiguity in the two trees represents reticulation evolution (eg. hybridization): the trees indicate that the LCA of taxa B and C evolved from the LCA of A to C and the LCA of A to D, but the LCA of A to D is not ancestral to the LCA of A to C. This, however, appears quite strange from an evolutionary point of view. A simple explanation for the conflict between the two trees would be that D is a hybrid of the lineage leading to A, which is the sister of B + C, and E.
 |
| A simple evolutionary scenario explaining the difference between the two conflicting trees: A is the paternal, and E the maternal donor of the hybrid D. |
As shown in tis figure, the LCA of A to D equals the LCA of A to C (depicted as two different nodes in the EUN), and the LCA of A+D and D+E are just (the precursors of) A and E . Taxon D is related to the ABC clade because the paternal donor has been A, plus to the E lineage via its mother.
This leads us to a principal question: do we want to reduce CUNs, which are splits graphs depicting all splits in a set of trees, ie. competing topological alternatives, to directed phylogenetic networks at all? The EUN has fewer edges (and nodes) than the CUN, but it still is an overly complex graph even for potentially very simple evolutionary scenarios.
On the Mesquite discussion group, a question was asked whether EUNs should be implemented as a means to quickly investigate conflict between trees. The answer to that question is: no. Consensus networks (CUNs) will be more than sufficient, since they are splits-based not node-based.
One application of EUNs may be ancestral state reconstruction. Character progression could be modeled the same way as it currently is along trees. Instead of viewing the nodes as actual LCAs in a reticulation scenario, one could consider them as competing alternative LCAs, and use the results of the ancestral state reconstruction along the EUN, to make a choice among alternatives, or simply to compare different evolutionary scenarios in the same graph.


No comments:
Post a Comment